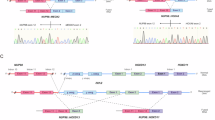
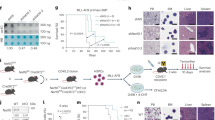
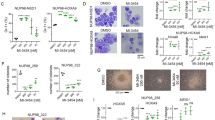
Abstract
NUP98 rearrangements associated with acute myeloid leukemia and myelodysplastic syndromes generate NUP98-fusion proteins. One such fusion protein, NUP98::DDX10, contains the putative RNA helicase DDX10. The molecular mechanism by which NUP98::DDX10 induces leukemia is not well understood. Here, we show that 24 amino acids within the DDX10 moiety of NUP98::DDX10 are crucial for cell immortalization and leukemogenesis. NOL10, nucleolar protein 10, interacts with the 24 amino acids, and NOL10 is a critical dependency of NUP98::DDX10 leukemia development. Studies in a mouse model of NUP98::DDX10 leukemia showed that loss of Nol10 impaired disease progression and improved survival. We also identified a novel function of NOL10 in that it acts cooperatively with NUP98::DDX10 to regulate serine biosynthesis pathways and stabilize ATF4 mRNA. Collectively, these findings suggest that NOL10 is a critical regulator of NUP98::DDX10 leukemia and that targeting NOL10 (or the serine synthesis pathway regulated by NOL10) may be an effective therapeutic approach.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501.
Nakamura T, Largaespada D, Lee M, Johnson L, Ohyashiki K, Toyama K, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–8.
Brown J, Jawad M, Twigg SRF, Saracoglu K, Sauerbrey A, Thomas AE, et al. A cryptic t(5;11)(q35;p15.5) in 2 children with acute myeloid leukemia with apparently normal karyotypes, identified by a multiplex fluorescence in situ hybridization telomere assay. Blood. 2002;99:2526–31.
Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, et al. The inv(11)(p15q22) chromosome translocation of de novo and therapy- related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–44.
Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood. 2011;118:6247–57.
Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021;595:591–5.
Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, et al. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell. 2016;30:863–78.
Shima Y, Yumoto M, Katsumoto T, Kitabayashi I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia. 2017;31:2200–10.
Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12.
Bammert L, Jonas S, Ungricht R, Kutay U. Human AATF/Che-1 forms a nucleolar protein complex with NGDN and NOL10 required for 40S ribosomal subunit synthesis. Nucleic Acids Res. 2016;44:9803–20.
Jin X, Tanaka H, Jin M, Fujita K, Homma H, Inotsume M, et al. PQBP5/NOL10 maintains and anchors the nucleolus under physiological and osmotic stress conditions. Nat Commun. 2023;14:1–5.
Shima Y, Honma Y, Kitabayashi I. PML-RARα and its phosphorylation regulate PML oligomerization and HIPK2 stability. Cancer Res. 2013;73:4278–88.
Xu S, Powers MA. Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis. Mol Biol Cell. 2010;21:1585–96.
Schmoellerl J, Barbosa IAM, Eder T, Brandstoetter T, Schmidt L, Maurer B, et al. CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood. 2020;136:387–400.
Jevtic Z, Matafora V, Casagrande C, Santoro F, Minucci S, Garré M, et al. SMARCA5 interacts with NUP98-NSD1 oncofusion protein and sustains hematopoietic cells transformation. J Exp Clin Cancer Res. 2022;41:34.
Terlecki-Zaniewicz S, Humer T, Eder T, Schmoellerl J, Heyes E, Manhart G, et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat Struc Mol Biol. 2021;28:190–201.
DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–81.
Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–62.
Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–57.
Wang B, Zhang L, Dai T, Qin Z, Lu H, Zhang L, et al. Liquid–liquid phase separation in human health and diseases. Signal Transduct Target Ther. 2021;6:290.
Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–55.
Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–33.
Takahashi Y, Lallemand-Breitenbach V, Zhu J, de Thé H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–24.
Nguyen LA, Pandolfi PP, Aikawa Y, Tagata Y, Ohki M, Kitabayashi I. Physical and functional link of the leukemia-associated factors AML1 and PML. Blood. 2005;105:292–300.
Davis JL, Fallon HJ, Morris HP. Two enzymes of serine metabolism in rat liver and hepatomas. Cancer Res. 1970;30:2917–20.
Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzym Regul. 1984;22:325–400.
Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–30.
Liu J, Guo S, Li Q, Yang L, Xia Z, Zhang L, et al. Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J Neurooncol. 2013;111:245–55.
Tajan M, Hennequart M, Cheung EC, Zani F, Hock AK, Legrave N, et al. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat Commun. 2021;12:1–16.
Allen RW, Moskowitz M. Arrest of cell growth in the G1 phase of the cell cycle by serine deprivation. Exp Cell Res. 1978;116:127–37.
Rowe PB, Sauer D, Fahey D, Craig G, McCairns E. One-carbon metabolism in lectin-activated human lymphocytes. Arch Biochem Biophys. 1985;236:277–88.
Davis SR, Stacpoole PW, Williamson J, Kick LS, Quinlivan EP, Coats BS, et al. Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab. 2004;286:272–9.
Acknowledgements
This study was supported by Project for Promotion of Cancer Research and Therapeutic Evolution (P-PROMOTE) from AMED.
Author information
Authors and Affiliations
Contributions
YS and IK conceived and designed the experiments. YS, KY, YK, KS, and YA performed the experiments. YS and KY performed the bioinformatic analysis. YS, KY, YK, KS, and IK analyzed the data. YS, KS, and IK wrote the manuscript. All authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shima, Y., Yamagata, K., Kuroki, Y. et al. Loss of NOL10 leads to impaired disease progression of NUP98::DDX10 leukemia. Leukemia 39, 1368–1379 (2025). https://doi.org/10.1038/s41375-025-02607-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41375-025-02607-5