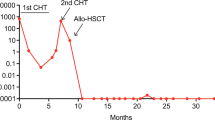
Abstract
NUP98 rearrangements represent a distinct, high-risk subtype of acute myeloid leukemia (AML), particularly in pediatric patients. However, their prevalence, genetic features, and clinical implications in adult AML remain poorly characterized. Using targeted-capture sequencing, we identified 41 cases with NUP98 rearrangements among 1569 AML cases, representing the majority of 44 NUP98-rearranged cases detected across 4753 myeloid neoplasms. Fifteen distinct fusion partners were detected, with NUP98::NSD1 and NUP98::HOXA9 being the most frequent. Notably, two novel fusions—NUP98::MEOX2 and NUP98::HOXA6—were identified. Co-mutations were relatively infrequent; FLT3-ITD and WT1 mutations were the most common, while NPM1 mutations were exclusive. FLT3-ITD was significantly enriched in NUP98::NSD1 cases, whereas TET2 mutations were more frequent in NUP98::HOXA9 cases. Clonal hierarchy analysis suggested that NUP98 rearrangements occur early in leukemogenesis. NUP98-rearranged AML exhibited higher relapse rates and shorter event-free survival. Specifically, NUP98::NSD1 was associated with a poor induction response, whereas NUP98::HOXA9 and NUP98 fusions with other partners showed higher remission rates but frequent relapse. Allogeneic hematopoietic stem cell transplantation was associated with better survival, underscoring its significance. These findings reveal the genetic and clinical heterogeneity of NUP98-rearranged AML in adults and support its classification as a distinct entity, highlighting the need for fusion partner-specific therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data generated in this study are available upon request from the corresponding author.
References
Bolouri H, Farrar JE, Triche T Jr, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–12.
Struski S, Lagarde S, Bories P, Puiseux C, Prade N, Cuccuini W, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31:565–72.
Bertrums EJM, Smith JL, Harmon L, Ries RE, Wang YJ, Alonzo TA, et al. Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia. Haematologica. 2023;108:2044–58.
Umeda M, Ma J, Westover T, Ni Y, Song G, Maciaszek JL, et al. A new genomic framework to categorize pediatric acute myeloid leukemia. Nat Genet. 2024;56:281–93.
Frerich C, Li P, Raess PW, Dunlap J, Lachowiez C, Solis-Ruiz J, et al. Landscape of somatic mutations and clonal evolution in NUP98-rearranged adult acute myeloid leukaemia. Br J Haematol. 2025;206:1097–102.
Tian J, Zhu Y, Li J, Yang G, Weng X, Huang T, et al. The landscape of NUP98 rearrangements clinical characteristics and treatment response from 1491 acute leukemia patients. Blood Cancer J. 2024;14:81.
Kim N, Choi YJ, Cho H, Jang JE, Lee ST, Song J, et al. NUP98 is rearranged in 5.0% of adult East Asian patients with AML. Blood Adv. 2024;8:5122–5.
Miyajima T, Onozawa M, Yoshida S, Miyashita N, Kimura H, Takahashi S, et al. Clinical implications of NUP98::NSD1 fusion at diagnosis in adult FLT3-ITD positive AML. Eur J Haematol. 2023;111:620–7.
Michmerhuizen NL, Klco JM, Mullighan CG. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood. 2020;136:2275–89.
Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021;595:591–5.
Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW, et al. Phase Separation Mediates NUP98 Fusion Oncoprotein Leukemic Transformation. Cancer Discov. 2022;12:1152–69.
Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98:1264–7.
Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–8.
Borrow J, Shearman AM, Stanton VP Jr, Becher R, Collins T, Williams AJ, et al. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–67.
Suzuki A, Ito Y, Sashida G, Honda S, Katagiri T, Fujino T, et al. t(7;11)(p15;p15) Chronic myeloid leukaemia developed into blastic transformation showing a novel NUP98/HOXA11 fusion. Br J Haematol. 2002;116:170–2.
Fujino T, Suzuki A, Ito Y, Ohyashiki K, Hatano Y, Miura I, et al. Single-translocation and double-chimeric transcripts: detection of NUP98-HOXA9 in myeloid leukemias with HOXA11 or HOXA13 breaks of the chromosomal translocation t(7;11)(p15;p15). Blood. 2002;99:1428–33.
Taketani T, Taki T, Shibuya N, Kikuchi A, Hanada R, Hayashi Y. Novel NUP98-HOXC11 fusion gene resulted from a chromosomal break within exon 1 of HOXC11 in acute myeloid leukemia with t(11;12)(p15;q13). Cancer Res. 2002;62:4571–4.
Panagopoulos I, Isaksson M, Billstrom R, Strombeck B, Mitelman F, Johansson B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13). Genes Chromosomes Cancer. 2003;36:107–12.
Su Z, Liu X, Zhang Y, Wang W, Li X, Yu J, et al. Transcriptional features of acute leukemia with promyelocytic differentiation lacking retinoic acid receptor rearrangements. Haematologica. 2023;108:3120–4.
Taketani T, Taki T, Shibuya N, Ito E, Kitazawa J, Terui K, et al. The HOXD11 gene is fused to the NUP98 gene in acute myeloid leukemia with t(2;11)(q31;p15). Cancer Res. 2002;62:33–7.
Cheng WY, Li JF, Zhu YM, Lin XJ, Wen LJ, Zhang F, et al. Transcriptome-based molecular subtypes and differentiation hierarchies improve the classification framework of acute myeloid leukemia. Proc Natl Acad Sci USA. 2022;119:e2211429119.
Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998;58:4269–73.
Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111:5672–82.
Soler G, Kaltenbach S, Dobbelstein S, Broccardo C, Radford I, Mozziconacci MJ, et al. Identification of GSX2 and AF10 as NUP98 partner genes in myeloid malignancies. Blood Cancer J. 2013;3:e124.
Nakamura T, Yamazaki Y, Hatano Y, Miura I. NUP98 is fused to PMX1 homeobox gene in human acute myelogenous leukemia with chromosome translocation t(1;11)(q23;p15). Blood. 1999;94:741–7.
Gervais C, Mauvieux L, Perrusson N, Helias C, Struski S, Leymarie V, et al. A new translocation t(9;11)(q34;p15) fuses NUP98 to a novel homeobox partner gene, PRRX2, in a therapy-related acute myeloid leukemia. Leukemia. 2005;19:145–8.
Lisboa S, Cerveira N, Bizarro S, Correia C, Vieira J, Torres L, et al. POU1F1 is a novel fusion partner of NUP98 in acute myeloid leukemia with t(3;11)(p11;p15). Mol Cancer. 2013;12:5.
Choi YJ, Lee S, Kim Y, Shin S, Lee KA. POU6F2, a novel fusion partner of NUP98 in acute myeloid leukaemia: A case report. Br J Haematol. 2024;205:1632–5.
Heald JS, Lopez AM, Pato ML, Ruiz-Xiville N, Cabezon M, Zamora L, et al. Identification of novel NUP98 fusion partners and comutations in acute myeloid leukemia: an adult cohort study. Blood Adv. 2024;8:2691–4.
Chou WC, Chen CY, Hou HA, Lin LI, Tang JL, Yao M, et al. Acute myeloid leukemia bearing t(7;11)(p15;p15) is a distinct cytogenetic entity with poor outcome and a distinct mutation profile: comparative analysis of 493 adult patients. Leukemia. 2009;23:1303–10.
Roussy M, Bilodeau M, Jouan L, Tibout P, Laramee L, Lemyre E, et al. NUP98-BPTF gene fusion identified in primary refractory acute megakaryoblastic leukemia of infancy. Genes Chromosomes Cancer. 2018;57:311–9.
Schwartz JR, Ma J, Lamprecht T, Walsh M, Wang S, Bryant V, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8:1557.
van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–46.
Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood. 2002;99:3857–60.
Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21:842–4.
Tembrink M, Gerding WM, Wieczorek S, Mika T, Schroers R, Nguyen HP, et al. Novel NUP98::ASH1L gene fusion in acute myeloid leukemia detected by optical genome mapping. Cancers. 2023;15.
Kaltenbach S, Soler G, Barin C, Gervais C, Bernard OA, Penard-Lacronique V, et al. NUP98-MLL fusion in human acute myeloblastic leukemia. Blood. 2010;116:2332–5.
Ikoma Y, Nakamura N, Kaneda Y, Takamori H, Seki T, Hiramoto N, et al. Impact of myelodysplasia-related gene mutations and residual mutations at remission in venetoclax/azacitidine for AML. Leukemia. 2025;39:1362–7.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7.
Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. N Engl J Med. 2015;373:35–47.
Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–68.
Ahuja HG, Felix CA, Aplan PD. The t(11;20)(p15;q11) chromosomal translocation associated with therapy-related myelodysplastic syndrome results in an NUP98-TOP1 fusion. Blood. 1999;94:3258–61.
Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, et al. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–44.
Gao L, Zhang F, Wen L, Wang Z, Ruan C, Chen S. Novel NUP98:TNRC18 fusion transcript in acute myeloid leukemia: a case report and literature review. Blood Sci. 2025;7:e00232.
Chonabayashi K, Yoshida Y, Kitawaki T, Nannya Y, Nakamura M, Oshima S, et al. Acute myeloid leukemia with a cryptic NUP98/PRRX2 rearrangement developing after low-dose methotrexate therapy for rheumatoid arthritis. Ann Hematol. 2019;98:2841–3.
Kim JC, Zuzarte PC, Murphy T, Chan-Seng-Yue M, Brown AMK, Krzyzanowski PM, et al. Cryptic genomic lesions in adverse-risk acute myeloid leukemia identified by integrated whole genome and transcriptome sequencing. Leukemia. 2020;34:306–11.
Ostronoff F, Othus M, Gerbing RB, Loken MR, Raimondi SC, Hirsch BA, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124:2400–7.
Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–56.
Kivioja JL, Thanasopoulou A, Kumar A, Kontro M, Yadav B, Majumder MM, et al. Dasatinib and navitoclax act synergistically to target NUP98-NSD1(+)/FLT3-ITD(+) acute myeloid leukemia. Leukemia. 2019;33:1360–72.
Acknowledgements
The authors acknowledge the patients who participated in this study and their families. The authors thank the clinical research staff and caregivers at all participating sites. In preparing this work, ChatGPT was used to proofread the manuscript.
Funding
This work was supported in part by grants from the Japan Agency for Medical Research and Development (JP19cm0106235h0002, JP22bm0804004h0006 (K.C. and Y.Y.), JP15cm0106056h0005, JP19cm0106501h0004, JP16ck0106073h0003, JP19ck0106250h0003 (S.O.), JP19ck0106353h0003 (Y.Nannya.)), Core Research for Evolutional Science and Technology (JP19gm1110011) (S.O.), and JSPS KAKENHI (JP21K08414 (K.C.), JP25H01055 (M.S.-Y)), a grant from the Kobayashi Foundation for Cancer Research (K.C.), and a grant from the International Joint Usage/Research Center, IMSUT (24-2103 (K.C. and Y.Nannya).
Author information
Authors and Affiliations
Contributions
KC, M.Iwasaki, JK, and Y.Nannya conceived and designed the study and wrote the manuscript. KC, M.Iwasaki, and Y.Nannya collected and curated the data. SO and Y.Nannya performed targeted-capture sequencing. HT, AY, SO, and Y.Nannya analyzed and interpreted the sequencing data. KC performed most of the experiments. T.Kawata, SM, and YY performed some experiments. MS, T.Kondo, TH, YU, AG, MW, SK, Y.I., HK, KI, KM, T.Kitano, YT, Y.Nakabou, NS, NK, TF, M.Ichikawa, YM, SF, MS-Y, and AT-K collected patient samples and data. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chonabayashi, K., Iwasaki, M., Kanda, J. et al. NUP98 rearrangements in adult AML patients: evaluation of clinical implications and identification of novel fusion partners. Leukemia (2026). https://doi.org/10.1038/s41375-025-02848-4
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41375-025-02848-4