Abstract
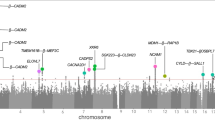
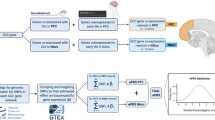
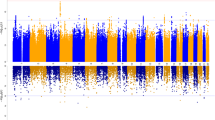
Genomic correlates of impulsivity have been identified in several genome-wide association studies (GWAS) using cross-sectional designs, but no studies have investigated the molecular genetic correlates of impulsivity phenotypes using longitudinally constructed traits. In 3860 unrelated European participants in the Avon Longitudinal Study of Parents and Children (ALSPAC), we constructed longitudinal phenotypes for delay discounting and impulsive personality traits (as measured by the UPPS-P impulsive behavior scales) via assessment at ages 24, 26, and 28. We conducted GWASs of impulsivity using both cross-sectional and longitudinal phenotypes, estimated heritability and their phenotypic and genetic correlations, and evaluated their association with recently-developed polygenic risk scores (PRSs) for the impulsivity indicators themselves and also related psychiatric conditions. Latent growth curve modeling revealed a stable intercept over time for all impulsivity phenotypes. High genetic correlation of cross-sectional measures over time suggested a stable genetic component for delay discounting (rg = 0.53–0.99) and sensation seeking (rg = 0.99). Heritability estimates of the stable longitudinal phenotypes substantively improved as compared to their cross-sectional counterparts, revealing a significant SNP-heritability for delay discounting (0.22; p = 0.03) and sensation seeking (0.35; p = 0.0007). Consistent with previous reports, GWAS and gene-based analyses revealed associations between specific longitudinal impulsivity indicators and CADM2 and NCAM1 genes. The PRSs for the impulsivity indicators and disorders related to self-regulation were also significantly associated with longitudinal impulsivity traits. Finally, we validated the associations between longitudinal impulsivity phenotypes and their PRSs in an independent 13-wave longitudinal study (n = 1019) and the benefit of longitudinal phenotypes in simulation studies. In this first longitudinal genetic study of impulsivity traits, the results revealed stable genomic correlates of delay discounting and sensation seeking over time and further validated the utility of recently-developed PRSs, both in relation to the observed traits and in connecting them to psychiatric disorders. More generally, these findings support using latent intercepts as novel longitudinal phenotypes to boost signal for heritability and genomic correlates of mechanisms contributing to psychiatric disease liability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Genotype and phenotype data are available through the ALSPAC data team pending a successful application. Access to the ALSPAC data was granted under project # B3136. Summary statistics generated in the current study, including a total of 6 LGC GWASs and 18 cross-sectional GWASs, will be made available on GWAS catalog (https://www.ebi.ac.uk/gwas).
References
Mackillop J. Integrating behavioral economics and behavioral genetics: delayed reward discounting as an endophenotype for addictive disorders. J Exp Anal Behav. 2013;99:14–31.
Cyders MA, Littlefield AK, Coffey S, Karyadi KA. Examination of a short English version of the UPPS-P impulsive behavior scale. Addict Behav. 2014;39:1372–6.
Patton JH, Stanford MS, Barratt ES. Factor structure of the barratt impulsiveness scale. J Clin Psychol. 1995;51:768–74.
De Wit H. Impulsivity as a determinant and consequence of drug use: a review of underlying processes. Addict Biol. 2009;14:22–31.
Bickel WK, Marsch LA. Toward a behavioral economic understanding of drug dependence: delay discounting processes. Addiction 2001;96:73–86.
Dick DM, Smith G, Olausson P, Mitchell SH, Leeman RF, O'Malley SS, et al. Understanding the construct of impulsivity and its relationship to alcohol use disorders. Addict Biol. 2010;15:217–26.
Amlung M, Vedelago L, Acker J, Balodis I, MacKillop J. Steep delay discounting and addictive behavior: a meta-analysis of continuous associations. Addiction 2017;112:51–62.
Murphy CM, Stojek MK, MacKillop J. Interrelationships among impulsive personality traits, food addiction, and body mass index. Appetite 2014;73:45–50.
MacKillop J, Amlung MT, Few LR, Ray LA, Sweet LH, Munafò MR. Delayed reward discounting and addictive behavior: a meta-analysis. Psychopharmacology. 2011;216:305–21.
Olson EA, Hooper CJ, Collins P, Luciana M. Adolescents’ performance on delay and probability discounting tasks: contributions of age, intelligence, executive functioning, and self-reported externalizing behavior. Pers Individ Dif. 2007;43:1886–97.
Beauchaine TP, Zisner AR, Sauder CL. Trait impulsivity and the externalizing spectrum. Annu Rev Clin Psychol. 2017;13:343–68.
Creswell KG, Wright AGC, Flory JD, Skrzynski CJ, Manuck SB. Multidimensional assessment of impulsivity-related measures in relation to externalizing behaviors. Psychol Med. 2019;49:1678–90.
MacKillop J, Weafer J, C Gray J, Oshri A, Palmer A, de Wit H. The latent structure of impulsivity: impulsive choice, impulsive action, and impulsive personality traits. Psychopharmacology. 2016;233:3361–70.
Gustavson DE, Friedman NP, Fontanillas P, Elson SL, Palmer AA, Sanchez-Roige S. The latent genetic structure of impulsivity and its relation to internalizing psychopathology. Psychol Sci. 2020;31:1025–35.
Brook DW, Brook JS, Zhang C, Whiteman M, Cohen P, Finch SJ. Developmental trajectories of cigarette smoking from adolescence to the early thirties: personality and behavioral risk factors. Nicotine Tob Res. 2008;10:1283–91.
Maggs JL, Schulenberg JE. Trajectories of alcohol use during the transition to adulthood. Alcohol Res. Health. 2005;28:195–201.
Green L, Myerson J, Ostaszewski P. Discounting of delayed rewards across the life span: age differences in individual discounting functions. Behav Process. 1999;46:89–96.
Green L, Myerson J, Lichtman D, Rosen S, Fry A. Temporal discounting in choice between delayed rewards: the role of age and income. Psychol Aging. 1996;11:79–84.
Sanchez-Roige S, Fontanillas P, Elson SL, Pandit A, Schmidt EM, Foerster JR, et al. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat Neurosci. 2018;21:16–18.
Sanchez-Roige S, Fontanillas P, Elson SL, Gray JC, De Wit H, MacKillop J, et al. Genome-wide association studies of impulsive personality traits (BIS-11 and UPPS-P) and drug experimentation in up to 22,861 adult research participants identify loci in the CACNA1I and CADM2 genes. J Neurosci. 2019;39:2562–72.
Sanchez-Roige S, Jennings MV, Thorpe HHA, Mallari JE, Van Der Werf LC, Bianchi SB, et al. CADM2 is implicated in impulsive personality and numerous other traits by genome- and phenome-wide association studies in humans and mice. Transl Psychiatry. 2023;13:167.
Littlefield AK, Sher KJ, Wood PK. Is ‘maturing out’ of problematic alcohol involvement related to personality change? J Abnorm Psychol. 2009;118:118–374.
Bergen SE, Gardner CO, Kendler KS. Age-related changes in heritability of behavioral phenotypes over adolescence and young adulthood: a meta-analysis. Twin Res Hum Genet. 2007;10:423–33.
Anokhin AP, Golosheykin S, Grant JD, Heath AC. Heritability of delay discounting in adolescence: a longitudinal twin study. Behav Genet. 2011;41:175–83.
Anokhin AP, Grant JD, Mulligan RC, Heath AC. The genetics of impulsivity: evidence for the heritability of delay discounting. Biol Psychiatry. 2015;77:887–94.
Adkins DE, Clark SL, Copeland WE, Kennedy M, Conway K, Angold A, et al. Genome-wide meta-analysis of longitudinal alcohol consumption across youth and early adulthood. Twin Res Hum Genet. 2015;18:335–47.
Dick DM, Cho SB, Latendresse SJ, Aliev F, Nurnberger JI, Edenberg HJ, et al. Genetic influences on alcohol use across stages of development: GABRA2 and longitudinal trajectories of drunkenness from adolescence to young adulthood. Addict Biol. 2014;19:1055–64.
Alves AC, De Silva NMG, Karhunen V, Sovio U, Das S, Rob Taal H, et al. GWAS on longitudinal growth traits reveals different genetic factors influencing infant, child, and adult BMI. Sci Adv. 2019;5:eaaw3095.
Vicuña L, Barrientos E, Norambuena T, Alvares D, Gana JC, Leiva-Yamaguchi V, et al. New insights from GWAS on BMI-related growth traits in a longitudinal cohort of admixed children with Native American and European ancestry. IScience. 2023;26:106091.
Amlung M, Petker T, Jackson J, Balodis I, Mackillop J. Steep discounting of delayed monetary and food rewards in obesity: a meta-analysis. Psychol Med. 2016;46:2423–34.
Ko S, German CA, Jensen A, Shen J, Wang A, Mehrotra DV, et al. GWAS of longitudinal trajectories at biobank scale. Am J Hum Genet. 2022;109:433–45.
McNeish D, Matta T. Differentiating between mixed-effects and latent-curve approaches to growth modeling. Behav Res Methods. 2018;50:1398–414.
Curran PJ, Obeidat K, Losardo D. Twelve frequently asked questions about growth curve modeling. J Cogn Dev. 2010;11:121–36.
Nurmi EL, Laughlin CP, de Wit H, Palmer AA, MacKillop J, Cannon TD, et al. Polygenic contributions to performance on the balloon analogue risk task. Mol Psychiatry. 2023;28:3524–30.
Fraser A, Macdonald-wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110.
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort profile: the ’Children of the 90s’—The index offspring of the avon longitudinal study of parents and children. Int J Epidemiol. 2013;42:11–27.
Northstone K, Lewcock M, Groom A, Boyd A, Macleod J, Timpson N, et al. The Avon longitudinal study of parents and children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51.
Kirby KN, Petry NM, Bickel WK. Heroin addicts have higher discount rates for delayed rewards than non-drug-using controls. J Exp Psychol Gen. 1999:128:78–87
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inf. 2009;42:377–81.
Joshua C., Gray Michael T., Amlung Abraham A., Palmer James, MacKillop. Syntax for calculation of discounting indices from the monetary choice questionnaire and probability discounting questionnaire. J Exp Anal Behav. 2016:106:156–63.
Koffarnus MN, Bickel WK. A 5-trial adjusting delay discounting task: accurate discount rates in less than one minute. Exp Clin Psychopharmacol. 2014;22:222–8.
Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53, 831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9
Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Deng WQ, Belisario K, Gray JC, Levitt EE, Mohammadi‐Shemirani P, Singh D, et al. Leveraging related health phenotypes for polygenic prediction of impulsive choice, impulsive action, and impulsive personality traits in 1534 European ancestry community adults. Genes Brain Behav. 2023;22:e12848.
Visscher PM, Hemani G, Vinkhuyzen AAE, Chen GB, Lee SH, Wray NR, et al. Statistical power to detect genetic (Co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269.
Rosseel Y Lavaan: an R package for structural equation modeling. J Stat Softw. 2012;48:1–36.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Consortium SWG of the PG, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Chen B, Craiu RV, Strug LJ, Sun L. The X factor: a robust and powerful approach to X-chromosome-inclusive whole-genome association studies. Genet Epidemiol. 2021;45:694–709.
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7.
R Core Team. R core team (2021). R: a language and environment for statistical computing R foundation for statistical computing, Vienna, Austria. URL: https://www.r-project.org. 2021.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, et al. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–30.
Karlsson Linnér R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24:1367–76.
Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–57.
Nagel M, Jansen PR, Stringer S, Watanabe K, De Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Hatoum AS, Colbert SMC, Johnson EC, Huggett SB, Deak JD, Pathak G, et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Nat Ment Health. 2023;1:210–23.
Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–18.
Saunders GRB, Wang X, Chen F, Jang SK, Liu M, Wang C, et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 2022;612:720–4.
Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41:469–80.
Quinn PD, Harden KP. Differential changes in impulsivity and sensation seeking and the escalation of substance use from adolescence to early adulthood. Dev Psychopathol. 2013;25:223–39.
Harden KP, Tucker-Drob EM. Individual differences in the development of sensation seeking and impulsivity during adolescence: further evidence for a dual systems model. Dev Psychol. 2011;47:739–46.
Dong Y, Peng CYJ. Principled missing data methods for researchers. Springerplus 2013;2:222.
Pasman JA, Chen Z, Smit DJA, Vink JM, Van Den Oever MC, Pattij T, et al. The CADM2 gene and behavior: a phenome-wide scan in UK-biobank. Behav Genet. 2022;52:306–14.
Sanchez-Roige S, Palmer AA, Clarke TK. Recent efforts to dissect the genetic basis of alcohol use and abuse. Biol Psychiatry. 2020;87:609–18.
Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke TK, Shirali M, et al. Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet. 2018;50:6–11.
Bierut LJ. Genetic vulnerability and susceptibility to substance dependence. Neuron 2011;69:618–27.
MacKillop J, Obasi EM, Amlung MT, McGeary JE, Knopik VS. The role of genetics in nicotine dependence: mapping the pathways from genome to syndrome. Curr Cardiovasc Risk Rep. 2010;4:446–53.
MacKillop J, Gray JC, Bidwell LC, Bickel WK, Sheffer CE, McGeary JE. Genetic influences on delay discounting in smokers: examination of a priori candidates and exploration of dopamine-related haplotypes. Psychopharmacology. 2015;232:3731–9.
Syan SK, González-Roz A, Amlung M, Sweet LH, MacKillop J. Delayed reward discounting as a prognostic factor for smoking cessation treatment outcome: a systematic review. Nicotine Tob Res. 2021;23:1636–45.
Rogerson O, Prudenzi A, O’Connor DB. Exploring the relationship between suicide vulnerability, impulsivity and executive functioning during COVID-19: a longitudinal analysis. Psychiatry Res Commun. 2022;2:100088.
Acknowledgements
We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We are especially thankful to all the staff and participants of the Population Assessment for Tomorrow’s Health registry conducted at St. Joseph’s Healthcare Hamilton/McMaster University. This research was enabled in part by support provided by SHARCNET (https://www.sharcnet.ca/) and the Digital Research Alliance of Canada (alliance can .ca). The PRS analyses included summary statistics from published GWASs made available by the Psychiatric Genomics Consortium (PGC), the Million Veterans Program (MVP), the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN), and 23andMe.
Funding
The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. Genomewide genotyping data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. A comprehensive list of grants funding is available on the ALSPAC website. This research was specifically funded by the Peter Boris Center for Addictions Research. JM is supported by the Peter Boris Chair in Addictions Research and a Canada Research Chair in Translational Addiction Research (JM; CRC-2020-00179). MRM is a member of the Medical Research Council (MRC) Integrative Epidemiology Unit at the University of Bristol (MC_UU_00032/07). This publication is the work of the authors and WQD will serve as guarantor for the contents of this paper.
Author information
Authors and Affiliations
Contributions
JM conceived, designed the study, and acquired funding; WQD and KB designed the statistical models, analyzed data, and produced visualizations; WQD designed and carried out the genomic analyses; KB carried out phenotypic analyses; JM and MRM acquired data; WQD, KB, and JM drafted the manuscript. All authors discussed results, critically reviewed the content, provided revisions of the manuscript for important intellectual content, and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
JM is a principal in Beam Diagnostics, Inc. and a consultant to Clairvoyant Therapeutics, Inc. No other authors have disclosures.
Ethics statement
Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Informed consent for the use of data collected via questionnaires and clinics was obtained from participants following the recommendations of the ALSPAC Ethics and Law Committee at the time. All PATH CANN participants underwent informed consent, and the registry was approved by the Hamilton Integrated Review Ethics Board (#1074).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, W.Q., Belisario, K., Munafò, M.R. et al. Longitudinal characterization of impulsivity phenotypes boosts signal for genomic correlates and heritability. Mol Psychiatry 30, 608–618 (2025). https://doi.org/10.1038/s41380-024-02704-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-024-02704-4