Abstract
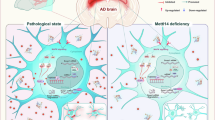
Alzheimer’s disease (AD) is the most common neurodegenerative disease, and diverse factors contribute to its pathogenesis. Previous studies have suggested the dysregulation of m6A modification involves in AD, but the underlying mechanism and targets remain largely unknown. In the present study, we have shown that the levels of Mettl3 and m6A modification are increased in specific brain regions of 5xFAD mice and post-mortem AD patients, respectively. Heterozygous deletion of neuronal Mettl3 (AD::Mettl3+/−) reduced Aβ plaques and inflammation, and improved learning and memory of AD mice, and vice versa for Mettl3 knock in (AD::Mettl3-KI). Mechanistically, we observed that the level of m6A modification of Lingo2 increased in 5xFAD mice and AD patients, which promoted the binding of Ythdf2 and enhanced the degradation of Lingo2 mRNA. The decreased level of Lingo2 promoted the interaction between APP and β-site amyloid precursor protein cleaving enzyme (Bace1), and subsequently enhanced Aβ production in AD mice, which can be inhibited by Mettl3 depletion. Both ectopic Lingo2 and the administration of Mettl3 inhibitor STM2457 significantly alleviated the neuropathology and behavioral deficits of AD mice. In summary, our study has revealed the important function of Mettl3 and m6A in the pathogenesis of AD and provided novel insight for the underlying mechanisms. Our study also suggests that m6A and Lingo2 could be potential therapeutic targets for AD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data have been deposited in GEO and the accession number is GSE268253.
Code availability
RNA-seq data have been deposited in GEO and the accession number is GSE268253.
References
Knopman DS, Amieva H, Petersen RC, Chetelat G, Holtzman DM, Hyman BT, et al. Alzheimer disease. Nat Rev Dis Primers. 2021;7:33.
Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1:15056.
Zhao J, Paganini L, Mucke L, Gordon M, Refolo L, Carman M, et al. Beta-secretase processing of the beta-amyloid precursor protein in transgenic mice is efficient in neurons but inefficient in astrocytes. J Biol Chem. 1996;271:31407–11.
Calhoun ME, Burgermeister P, Phinney AL, Stalder M, Tolnay M, Wiederhold KH, et al. Neuronal overexpression of mutant amyloid precursor protein results in prominent deposition of cerebrovascular amyloid. Proc Natl Acad Sci USA. 1999;96:14088–93.
Wang H, Kulas JA, Wang C, Holtzman DM, Ferris HA, Hansen SB. Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc Natl Acad Sci USA. 2021;118:e2102191118.
van Dyck CH, Swanson CJ, Aisen P, Bateman RJ, Chen C, Gee M, et al. Lecanemab in early Alzheimer’s disease. N Engl J Med. 2023;388:9–21.
Sims JR, Zimmer JA, Evans CD, Lu M, Ardayfio P, Sparks J, et al. Donanemab in early symptomatic Alzheimer disease: The TRAILBLAZER-ALZ 2 randomized clinical trial. JAMA. 2023;330:512–27.
Algamal M, Russ AN, Miller MR, Hou SS, Maci M, Munting LP, et al. Reduced excitatory neuron activity and interneuron-type-specific deficits in a mouse model of Alzheimer’s disease. Commun Biol. 2022;5:1323.
Busche MA, Eichhoff G, Adelsberger H, Abramowski D, Wiederhold KH, Haass C, et al. Clusters of hyperactive neurons near amyloid plaques in a mouse model of Alzheimer’s disease. Science. 2008;321:1686–9.
Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, et al. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron. 2007;55:697–711.
Zhang W, Xiong BR, Zhang LQ, Huang X, Yuan X, Tian YK, et al. The role of the GABAergic system in diseases of the central nervous system. Neuroscience. 2021;470:88–99.
Melgosa-Ecenarro L, Doostdar N, Radulescu CI, Jackson JS, Barnes SJ. Pinpointing the locus of GABAergic vulnerability in Alzheimer’s disease. Semin Cell Dev Biol. 2023;139:35–54.
Nuriel T, Angulo SL, Khan U, Ashok A, Chen Q, Figueroa HY, et al. Neuronal hyperactivity due to loss of inhibitory tone in APOE4 mice lacking Alzheimer’s disease-like pathology. Nat Commun. 2017;8:1464.
Boulias K, Greer EL. Biological roles of adenine methylation in RNA. Nat Rev Genet. 2023;24:143–60.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21:36–51.
Xiong X, James BT, Boix CA, Park YP, Galani K, Victor MB, et al. Epigenomic dissection of Alzheimer’s disease pinpoints causal variants and reveals epigenome erosion. Cell. 2023;186:4422–37.e4421.
Berson A, Nativio R, Berger SL, Bonini NM. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41:587–98.
Nativio R, Lan Y, Donahue G, Sidoli S, Berson A, Srinivasan AR, et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat Genet. 2020;52:1024–35.
Gao C, Shen X, Tan Y, Chen S. Pathogenesis, therapeutic strategies and biomarker development based on “omics” analysis related to microglia in Alzheimer’s disease. J Neuroinflammation. 2022;19:215.
Castro-Hernandez R, Berulava T, Metelova M, Epple R, Pena Centeno T, Richter J, et al. Conserved reduction of m(6)A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts. Proc Natl Acad Sci USA. 2023;120:e2204933120.
Han M, Liu Z, Xu Y, Liu X, Wang D, Li F, et al. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front Neurosci. 2020;14:98.
Yin H, Ju Z, Zheng M, Zhang X, Zuo W, Wang Y, et al. Loss of the m6A methyltransferase METTL3 in monocyte-derived macrophages ameliorates Alzheimer’s disease pathology in mice. PLoS Biol. 2023;21:e3002017.
Huang H, Camats-Perna J, Medeiros R, Anggono V, Widagdo J. Altered expression of the m6A methyltransferase METTL3 in Alzheimer’s disease. eNeuro 2020;7:ENEURO.0125-20.2020.
Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22:17.
Zhao F, Xu Y, Gao S, Qin L, Austria Q, Siedlak SL, et al. METTL3-dependent RNA m(6)A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol Neurodegener. 2021;16:70.
Liu Z, Xia Q, Zhao X, Zheng F, Xiao J, Ge F, et al. The landscape of m6A regulators in multiple brain regions of Alzheimer’s disease. Mol Neurobiol. 2023;60:5184–98.
Jiang L, Lin W, Zhang C, Ash PEA, Verma M, Kwan J, et al. Interaction of tau with HNRNPA2B1 and N(6)-methyladenosine RNA mediates the progression of tauopathy. Mol Cell. 2021;81:4209–4227.e4212.
Jiang L, Roberts R, Wong M, Zhang L, Webber CJ, Libera J, et al. beta-amyloid accumulation enhances microtubule associated protein tau pathology in an APP(NL-G-F)/MAPT(P301S) mouse model of Alzheimer’s disease. Front Neurosci. 2024;18:1372297.
Tang Z, Cao J, Yao J, Fan X, Zhao J, Zhao M, et al. KDM1A-mediated upregulation of METTL3 ameliorates Alzheimer’s disease via enhancing autophagic clearance of p-Tau through m6A-dependent regulation of STUB1. Free Radic Biol Med. 2023;195:343–58.
Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013;5:48.
Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91.
Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36:177–88.
Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-Containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507.e498.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Tan J, Evin G. Β-site APP-cleaving enzyme 1 trafficking and Alzheimer’s disease pathogenesis. J Neurochemistry. 2012;120:869–80.
Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, et al. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat Med. 1995;1:1291–6.
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597–601.
Wu YW, Prakash KM, Rong TY, Li HH, Xiao Q, Tan LC, et al. Lingo2 variants associated with essential tremor and Parkinson’s disease. Hum Genet. 2011;129:611–5.
Vilarino-Guell C, Wider C, Ross OA, Jasinska-Myga B, Kachergus J, Cobb SA, et al. LINGO1 and LINGO2 variants are associated with essential tremor and Parkinson disease. Neurogenetics. 2010;11:401–8.
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–96.
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10.
Li P, Marshall L, Oh G, Jakubowski JL, Groot D, He Y, et al. Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat Commun. 2019;10:2246.
Chopra N, Wang R, Maloney B, Nho K, Beck JS, Pourshafie N, et al. MicroRNA-298 reduces levels of human amyloid-beta precursor protein (APP), beta-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol Psychiatry. 2021;26:5636–57.
Zhang Z, Li XG, Wang ZH, Song M, Yu SP, Kang SS, et al. delta-Secretase-cleaved Tau stimulates Abeta production via upregulating STAT1-BACE1 signaling in Alzheimer’s disease. Mol Psychiatry. 2021;26:586–603.
Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–2.
Hampel H, Hardy J, Blennow K, Chen C, Perry G, Kim SH, et al. The Amyloid-beta pathway in Alzheimer’s disease. Mol Psychiatry. 2021;26:5481–503.
De Strooper B, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol. 2010;6:99–107.
Chen K, Nan J, Xiong X. Genetic regulation of m(6)A RNA methylation and its contribution in human complex diseases. Sci China Life Sci. 2024.
Li L, Xia X, Yang T, Sun Y, Liu X, Xu W, et al. RNA methylation: a potential therapeutic target in autoimmune disease. Int Rev Immunol. 2024;43:160–77.
Wang C, Hou X, Guan Q, Zhou H, Zhou L, Liu L, et al. RNA modification in cardiovascular disease: implications for therapeutic interventions. Signal Transduct Target Ther. 2023;8:412.
Yang L, Tian S, Zheng X, Zhang M, Zhou X, Shang Y, et al. N6-methyladenosine RNA methylation in liver diseases: from mechanism to treatment. J Gastroenterol. 2023;58:718–33.
Pomaville MM, He C. Advances in targeting RNA modifications for anticancer therapy. Trends Cancer. 2023;9:528–42.
Li C, Xu P, Huang Y, Wang Y, Wu Y, Li H, et al. RNA methylations in depression, from pathological mechanism to therapeutic potential. Biochem Pharmacol. 2023;215:115750.
Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m(6)A-YTHDF2-dependent manner. Oncogene. 2022;41:1622–33.
Zhang ZW, Teng X, Zhao F, Ma C, Zhang J, Xiao LF, et al. METTL3 regulates m(6)A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma. Cell Rep. 2022;41:111530.
Gao J, Fang Y, Chen J, Tang Z, Tian M, Jiang X, et al. Methyltransferase like 3 inhibition limits intrahepatic cholangiocarcinoma metabolic reprogramming and potentiates the efficacy of chemotherapy. Oncogene. 2023;42:2507–20.
Zhang Z, Wang M, Xie D, Huang Z, Zhang L, Yang Y, et al. METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018;28:1050–61.
Dong X, Shu L, Zhang J, Yang X, Cheng X, Zhao X, et al. Ogt-mediated O-GlcNAcylation inhibits astrocytes activation through modulating NF-kappaB signaling pathway. J Neuroinflammation. 2023;20:146.
Cheng J, Dong Y, Ma J, Pan R, Liao Y, Kong X, et al. Microglial Calhm2 regulates neuroinflammation and contributes to Alzheimer’s disease pathology. Sci Adv. 2021;7:eabe3600.
Mahan TE, Wang C, Bao X, Choudhury A, Ulrich JD, Holtzman DM. Selective reduction of astrocyte apoE3 and apoE4 strongly reduces Abeta accumulation and plaque-related pathology in a mouse model of amyloidosis. Mol Neurodegener. 2022;17:13.
Wang W, Zhao X, Shao Y, Duan X, Wang Y, Li J, et al. Mutation-induced DNMT1 cleavage drives neurodegenerative disease. Sci Adv. 2021;7:eabe8511.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–6.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660–1677.e1610.
Lo AC, Haass C, Wagner SL, Teplow DB, Sisodia SS. Metabolism of the “Swedish” amyloid precursor protein variant in Madin-Darby canine kidney cells. J Biol Chem. 1994;269:30966–73.
Acknowledgements
This work was supported in part by the Natural Science Foundation of Zhejiang Province (LD25H090001 to X.L.) and the National Key Research and Development Program of China (2017YFE0196600 to X.L.), and the Youth Innovation Promotion Association of CAS (Y2022040 to Y.Y.). We wish to thank the National Health and Disease Human Brain Tissue Resource Center for providing brain materials.
Author information
Authors and Affiliations
Contributions
XL conceptualized the project. XZ and QS performed immunostaining, qRT-PCR, western blots and immunoprecipitation with the help of XH and ZL CM, YC, YY and WQ analyzed MeRIP-seq data. XZ and A B analyzed human samples. XL, YY and BS discussed the data analysis. XL wrote the manuscript with the help of XZ, YY and BS.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, X., Ma, C., Sun, Q. et al. Mettl3 regulates the pathogenesis of Alzheimer’s disease via fine-tuning Lingo2. Mol Psychiatry 30, 4047–4063 (2025). https://doi.org/10.1038/s41380-025-02984-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-02984-4