Abstract
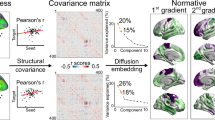
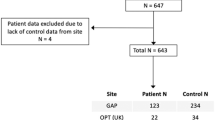
Pathological disturbances in schizophrenia have been suggested to propagate via the functional and structural connectome across the lifespan. However, how the connectome guides early cortical reorganization of developing schizophrenia remains unknown. Here, we used early-onset schizophrenia (EOS) as a neurodevelopmental disease model to investigate putative early pathologic origins propagating through the functional and structural connectome. We compared 95 patients with antipsychotic-naïve first-episode EOS and 99 typically developing controls (total n = 194; 120 females; 7–17 years of age). While patients showed widespread cortical thickness reductions, thickness increases were observed in primary cortical areas. Using normative connectomics models, we found that epicenters of thickness reductions were located in association regions linked to language, affective, and cognitive functions, while epicenters of thickness increases in EOS were located in sensorimotor regions subserving visual, somatosensory, and motor functions. Using post-mortem transcriptomic data of six donors, we observed that the epicenter map differentiated oligodendrocyte-related transcriptional changes at its sensory apex, whereas the association end was related to the expression of excitatory/inhibitory neurons. More generally, the epicenter map was associated with dysregulation of neurodevelopmental disorder genes and human accelerated region genes, suggesting potential common genetic determinants across diverse neurodevelopmental conditions. Taken together, our results highlight the developmentally rooted pathological origins of schizophrenia and its transcriptomic overlap with other neurodevelopmental disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Functional and structural disease epicenter maps of EOS and other data supporting our findings are available at https://github.com/Yun-Shuang/Neurodevelopmentally-rooted-epicenters-in-schizophrenia. The neurodevelopmental axis ranking data are available at https://github.com/PennLINC/S-A_ArchetypalAxis. Human gene expression data are available at https://human.brain-map.org/. Additional information can be made available upon reasonable request to the authors.
Code availability
Custom code was made publicly available under https://github.com/Yun-Shuang/Neurodevelopmentally-rooted-epicenters-in-schizophrenia. Epicenter calculation was based on ENIGMA Toolbox (https://enigma-toolbox.readthedocs.io/en/latest/); cognitive meta-analysis code was adapted from https://github.com/CNG-LAB/cngopen/blob/main/transdiagnostic_gradients/Scripts/Hettwer2022_Figure2_Transdiagnostic_Gradients.m; gene expression analyses were performed by the abagen toolbox (https://abagen.readthedocs.io/), combined with code under https://github.com/netneurolab/hansen_genescognition; gene enrichment analyses by metascape (https://metascape.org/gp/index.html#/main/step1); statistically analyses were carried out by BrainStat (https://github.com/MICA-MNI/BrainStat); visualizations were based on workbench (https://www.humanconnectome.org/software/connectome-workbench) and ggseg (https://ggseg.github.io/ggseg/), combined with ColorBrewer (https://github.com/scottclowe/cbrewer2).
References
Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–93.
Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18:727–40.
Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry. 2012;17:1228–38.
Friedrichs-Maeder CL, Griffa A, Schneider J, Huppi PS, Truttmann A, Hagmann P. Exploring the role of white matter connectivity in cortex maturation. PLoS One. 2017;12:e0177466.
Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–72.
Shafiei G, Markello RD, Makowski C, Talpalaru A, Kirschner M, Devenyi GA, et al. Spatial patterning of tissue volume loss in schizophrenia reflects brain network architecture. Biol Psychiatry. 2020;87:727–35.
Georgiadis F, Lariviere S, Glahn D, Hong E, Kochunov P, Mowry B, et al. Connectome architecture shapes large-scale cortical reorganization in schizophrenia: a worldwide ENIGMA study. Mol Psychiatry. 2024;29:1869–81.
Chopra S, Segal A, Oldham S, Holmes A, Sabaroedin K, Orchard ER, et al. Network-based spreading of gray matter changes across different stages of psychosis. JAMA Psychiatry. 2023;80:1246–57.
Godwin D, Alpert KI, Wang L, Mamah D. Regional cortical thinning in young adults with schizophrenia but not psychotic or non-psychotic bipolar I disorder. Int J Bipolar Disord. 2018;6:16.
Dukart J, Smieskova R, Harrisberger F, Lenz C, Schmidt A, Walter A, et al. Age-related brain structural alterations as an intermediate phenotype of psychosis. J Psychiatry Neurosci. 2017;42:307–19.
Palaniyappan L, Das TK, Winmill L, Hough M, James A. Progressive post-onset reorganisation of MRI-derived cortical thickness in adolescents with schizophrenia. Schizophr Res. 2019;208:477–8.
Voets NL, Hough MG, Douaud G, Matthews PM, James A, Winmill L, et al. Evidence for abnormalities of cortical development in adolescent-onset schizophrenia. Neuroimage. 2008;43:665–75.
Thormodsen R, Rimol LM, Tamnes CK, Juuhl-Langseth M, Holmen A, Emblem KE, et al. Age-related cortical thickness differences in adolescents with early-onset schizophrenia compared with healthy adolescents. Psychiatry Res. 2013;214:190–6.
Palaniyappan L, Hodgson O, Balain V, Iwabuchi S, Gowland P, Liddle P. Structural covariance and cortical reorganisation in schizophrenia: a MRI-based morphometric study. Psychol Med. 2019;49:412–20.
Wannan CMJ, Cropley VL, Chakravarty MM, Bousman C, Ganella EP, Bruggemann JM, et al. Evidence for network-based cortical thickness reductions in schizophrenia. Am J Psychiatry. 2019;176:552–63.
van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2016;21:547–53.
Lariviere S, Rodriguez-Cruces R, Royer J, Caligiuri ME, Gambardella A, Concha L, et al. Network-based atrophy modeling in the common epilepsies: a worldwide ENIGMA study. Sci Adv. 2020;6:eabc6457.
Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52.
Zeighami Y, Ulla M, Iturria-Medina Y, Dadar M, Zhang Y, Larcher KM, et al. Network structure of brain atrophy in de novo Parkinson’s disease. eLife. 2015;4:e08440.
Yau Y, Zeighami Y, Baker TE, Larcher K, Vainik U, Dadar M, et al. Network connectivity determines cortical thinning in early Parkinson’s disease progression. Nat Commun. 2018;9:12.
Hettwer MD, Lariviere S, Park BY, van den Heuvel OA, Schmaal L, Andreassen OA, et al. Coordinated cortical thickness alterations across six neurodevelopmental and psychiatric disorders. Nat Commun. 2022;13:6851.
Sydnor VJ, Larsen B, Bassett DS, Alexander-Bloch A, Fair DA, Liston C, et al. Neurodevelopment of the association cortices: patterns, mechanisms, and implications for psychopathology. Neuron. 2021;109:2820–46.
Chen CH, Fiecas M, Gutierrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, et al. Genetic topography of brain morphology. Proc Natl Acad Sci USA. 2013;110:17089–94.
Schmitt A, Falkai P, Papiol S. Neurodevelopmental disturbances in schizophrenia: evidence from genetic and environmental factors. J Neural Transm. 2023;130:195–205.
Owen MJ, O’Donovan MC. Schizophrenia and the neurodevelopmental continuum:evidence from genomics. World Psychiatry. 2017;16:227–35.
Rees E, Creeth HDJ, Hwu HG, Chen WJ, Tsuang M, Glatt SJ, et al. Schizophrenia, autism spectrum disorders and developmental disorders share specific disruptive coding mutations. Nat Commun. 2021;12:5353.
Crow TJ. Schizophrenia as the price that homo sapiens pays for language: a resolution of the central paradox in the origin of the species. Brain Res Brain Res Rev. 2000;31:118–29.
Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168.
Guardiola-Ripoll M, Fatjo-Vilas M. A systematic review of the human accelerated regions in schizophrenia and related disorders: where the evolutionary and neurodevelopmental hypotheses converge. Int J Mol Sci. 2023;24:3597.
Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832–44.
Fornito A, Arnatkeviciute A, Fulcher BD. Bridging the gap between connectome and transcriptome. Trends Cogn Sci. 2019;23:34–50.
Seidlitz J, Nadig A, Liu S, Bethlehem RAI, Vertes PE, Morgan SE, et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat Commun. 2020;11:3358.
Hansen JY, Shafiei G, Vogel JW, Smart K, Bearden CE, Hoogman M, et al. Local molecular and global connectomic contributions to cross-disorder cortical abnormalities. Nat Commun. 2022;13:4682.
Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7.
Wei Y, de Lange SC, Scholtens LH, Watanabe K, Ardesch DJ, Jansen PR, et al. Genetic mapping and evolutionary analysis of human-expanded cognitive networks. Nat Commun. 2019;10:4839.
Fan YS, Xu Y, Bayrak S, Shine JM, Wan B, Li H, et al. Macroscale thalamic functional organization disturbances and underlying core cytoarchitecture in early-onset schizophrenia. Schizophr Bull. 2023;49:1375–86.
Lariviere S, Paquola C, Park BY, Royer J, Wang Y, Benkarim O, et al. The ENIGMA toolbox: multiscale neural contextualization of multisite neuroimaging datasets. Nat Methods. 2021;18:698–700.
Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, et al. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage. 2018;178:540–51.
Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28:3095–114.
Markello RD, Arnatkeviciute A, Poline JB, Fulcher BD, Fornito A, Misic B. Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife. 2021;10:e72129.
Krishnan A, Williams LJ, McIntosh AR, Abdi H. Partial least squares (PLS) methods for neuroimaging: a tutorial and review. Neuroimage. 2011;56:455–75.
Hansen JY, Markello RD, Vogel JW, Seidlitz J, Bzdok D, Misic B. Mapping gene transcription and neurocognition across human neocortex. Nat Hum Behav. 2021;5:1240–50.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, et al. The human connectome project: a data acquisition perspective. Neuroimage. 2012;62:2222–31.
Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–65.
Keller AS, Sydnor VJ, Pines A, Fair DA, Bassett DS, Satterthwaite TD. Hierarchical functional system development supports executive function. Trends Cogn Sci. 2023;27:160–74.
Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–70.
Schwarzkopf DS, De Haas B, Rees G. Better ways to improve standards in brain-behavior correlation analysis. Front Hum Neurosci. 2012;6:200.
Abdi H. Partial least squares regression and projection on latent structure regression (PLS Regression). WIREs Comput Stat. 2010;2:97–106.
Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci USA. 2016;113:E469–478.
Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 2016;167:341–54. e312.
Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–56.
Zhao Y, Zhang Q, Shah C, Li Q, Sweeney JA, Li F, et al. Cortical thickness abnormalities at different stages of the illness course in schizophrenia: a systematic review and meta-analysis. JAMA Psychiatry. 2022;79:560–70.
van Erp TGM, Walton E, Hibar DP, Schmaal L, Jiang W, Glahn DC, et al. Cortical brain abnormalities in 4474 individuals with schizophrenia and 5098 control subjects via the enhancing neuro imaging genetics through meta analysis (ENIGMA) consortium. Biol Psychiatry. 2018;84:644–54.
Kirschner M, Paquola C, Khundrakpam BS, Vainik U, Bhutani N, Hodzic-Santor B, et al. Schizophrenia polygenic risk during typical development reflects multiscale cortical organization. Biol Psychiatry Glob Open Sci. 2023;3:1083–93.
Kirschner M, Hodzic-Santor B, Antoniades M, Nenadic I, Kircher T, Krug A, et al. Cortical and subcortical neuroanatomical signatures of schizotypy in 3004 individuals assessed in a worldwide ENIGMA study. Mol Psychiatry. 2022;27:1167–76.
van Haren NE, Schnack HG, Cahn W, van den Heuvel MP, Lepage C, Collins L, et al. Changes in cortical thickness during the course of illness in schizophrenia. Arch Gen Psychiatry. 2011;68:871–80.
Castellanos FX, Di Martino A, Craddock RC, Mehta AD, Milham MP. Clinical applications of the functional connectome. Neuroimage. 2013;80:527–40.
Valk SL, Xu T, Paquola C, Park BY, Bethlehem RAI, Vos de Wael R, et al. Genetic and phylogenetic uncoupling of structure and function in human transmodal cortex. Nat Commun. 2022;13:2341.
Baum GL, Cui Z, Roalf DR, Ciric R, Betzel RF, Larsen B, et al. Development of structure-function coupling in human brain networks during youth. Proc Natl Acad Sci USA. 2020;117:771–8.
Wang F, Lian C, Wu Z, Zhang H, Li T, Meng Y, et al. Developmental topography of cortical thickness during infancy. Proc Natl Acad Sci USA. 2019;116:15855–60.
Bethlehem RAI, Seidlitz J, White SR, Vogel JW, Anderson KM, Adamson C, et al. Brain charts for the human lifespan. Nature. 2022;604:525–33.
Gilmore JH, Knickmeyer RC, Gao W. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 2018;19:123–37.
Amlien IK, Fjell AM, Tamnes CK, Grydeland H, Krogsrud SK, Chaplin TA, et al. Organizing principles of human cortical development–thickness and area from 4 to 30 years: insights from comparative primate neuroanatomy. Cereb Cortex. 2016;26:257–67.
Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, et al. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci. 2008;28:3586–94.
Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. J Neurosci. 2004;24:8223–31.
Ball G, Seidlitz J, Beare R, Seal ML. Cortical remodelling in childhood is associated with genes enriched for neurodevelopmental disorders. Neuroimage. 2020;215:116803.
Kamholz J, Toffenetti J, Lazzarini RA. Organization and expression of the human myelin basic protein gene. J Neurosci Res. 1988;21:62–70.
Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, et al. Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci USA. 2012;109:16480–5.
Larsen B, Cui Z, Adebimpe A, Pines A, Alexander-Bloch A, Bertolero M, et al. A developmental reduction of the excitation:inhibition ratio in association cortex during adolescence. Sci Adv. 2022;8:eabj8750.
Toyoizumi T, Miyamoto H, Yazaki-Sugiyama Y, Atapour N, Hensch TK, Miller KD. A theory of the transition to critical period plasticity: inhibition selectively suppresses spontaneous activity. Neuron. 2013;80:51–63.
Sohal VS, Rubenstein JLR. Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol Psychiatry. 2019;24:1248–57.
Anticevic A, Murray JD. Rebalancing altered computations: considering the role of neural excitation and inhibition balance across the psychiatric spectrum. Biol Psychiatry. 2017;81:816–7.
Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009;1:102.
Capra JA, Erwin GD, McKinsey G, Rubenstein JL, Pollard KS. Many human accelerated regions are developmental enhancers. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130025.
Levchenko A, Kanapin A, Samsonova A, Gainetdinov RR. Human accelerated regions and other human-specific sequence variations in the context of evolution and their relevance for brain development. Genome Biol Evol. 2018;10:166–88.
Erady C, Amin K, Onilogbo T, Tomasik J, Jukes-Jones R, Umrania Y, et al. Novel open reading frames in human accelerated regions and transposable elements reveal new leads to understand schizophrenia and bipolar disorder. Mol Psychiatry. 2022;27:1455–68.
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362:eaat8464.
Acknowledgements
We are grateful to all the participants and their guardians in this study. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript. This work was supported by the National Natural Science Foundation of China (62403105, 62333003, 82121003, 62373079), the China Postdoctoral Science Foundation (2023M740524), Sichuan Province Innovative Talent Funding Project for Postdoctoral Fellows, Medical-Engineering Cooperation Funds from University of Electronic Science and Technology of China (ZYGX2021YGLH201), the Science and Technology Bureau of Chengdu Program (2024-YF05-00873-SN), the Youth Innovation Project of Sichuan Provincial Medical Association (Q23078), Medical Research Project of Chengdu (2024150). S.L.V. was also funded in part by Helmholtz Association’s Initiative and Networking Fund under the Helmholtz International Lab grant agreement InterLabs-0015, and the Canada First Research Excellence Fund (CFREF Competition 2, 2015–2016) awarded to the Healthy Brains, Healthy Lives initiative at McGill University, through the Helmholtz International BigBrain Analytics and Learning Laboratory (HIBALL). M.D.H. was funded by the Max Planck Society and the German Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Contributions
Y-SF, SLV and HC contributed to the conception and design of the work. YX, HC and MY contributed to the acquisition and interpretation of data for the work. Y-SF, PY, WS and CW contributed to the analysis of data. Y-SF wrote the manuscript. MDH, MK, and SLV revised it critically for important intellectual content. All authors reviewed initial drafts and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, YS., Xu, Y., Hettwer, M.D. et al. Neurodevelopmentally rooted epicenters in schizophrenia: sensorimotor-association spatial axis of cortical thickness alterations. Mol Psychiatry 31, 929–940 (2026). https://doi.org/10.1038/s41380-025-03193-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-03193-9