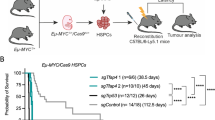
Abstract
Chromatin remodeling factors contribute to establish aberrant gene expression programs in cancer cells and therefore represent valuable targets for therapeutic intervention. BPTF (Bromodomain PhD Transcription Factor), a core subunit of the nucleosome remodeling factor (NURF), modulates c-MYC oncogenic activity in pancreatic cancer. Here, we analyze the role of BPTF in c-MYC-driven B-cell lymphomagenesis using the Eμ-Myc transgenic mouse model of aggressive B-cell lymphoma. We find that BPTF is required for normal B-cell differentiation without evidence of haploinsufficiency. In contrast, deletion of one Bptf allele is sufficient to delay lymphomagenesis in Eμ-Myc mice. Tumors arising in a Bptf heterozygous background display decreased c-MYC levels and pathway activity, together with increased activation of the NF-κB pathway, a molecular signature characteristic of human diffuse large B-cell lymphoma (DLBCL). In human B-cell lymphoma samples, we find a strong correlation between BPTF and c-MYC mRNA and protein levels, together with an anti-correlation between BPTF and NF-κB pathway activity. Our results indicate that BPTF is a relevant therapeutic target in B-cell lymphomas and that, upon its inhibition, cells acquire distinct oncogenic dependencies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol. 2013;14:r106.
Malysheva V, Mendoza-Parra MA, Saleem MA, Gronemeyer H. Reconstruction of gene regulatory networks reveals chromatin remodelers and key transcription factors in tumorigenesis. Genome Med. 2016;8:57.
Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–63.
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90.
Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–16.
Bishop PC, Rao VK, Wilson WH. Burkitt’s lymphoma: molecular pathogenesis and treatment. Cancer Investig. 2000;18:574–83.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11.
Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361.
Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303.
Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5.
Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706.
Dar AA, Majid S, Bezrookove V, Phan B, Ursu S, Nosrati M, et al. BPTF transduces MITF-driven prosurvival signals in melanoma cells. Proc Natl Acad Sci USA. 2016;113:6254–8.
Richart L, Carrillo-de Santa Pau E, Río-Machín A, de Andrés MP, Cigudosa JC, Lobo VJS. et al. Bptf is required for c-Myc transcriptional activity and in vivo tumorigenesis. Nat Commun. 2016;7:10153
Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vázquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–9.
Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8.
Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–71.
Mori S, Rempel RE, Chang JT, Yao G, Lagoo AS, Potti A, et al. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Eμ-myc model and human diffuse large B-cell lymphoma. Cancer Res. 2008;68:8525–34.
Rempel RE, Jiang X, Fullerton P, Tan TZ, Ye J, Lau JA, et al. Utilization of the Eμ-Myc mouse to model heterogeneity of therapeutic response. Mol Cancer Ther. 2014;13:3219–29.
Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–69.
Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–96.
Lee SC, Phipson B, Hyland CD, Leong HS, Allan RS, Lun A, et al. Polycomb repressive complex 2 (PRC2) suppresses Em-myc lymphoma. Blood. 2013;122:2654–63.
Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, et al. Essential role of chromatin remodelling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241.
Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–94.
Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–12.
Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92.
Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TFE, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–30.
Klapper W, Szczepanowski M, Burkhardt B, Berger H, Rosolowski M, Bentink S, et al. Molecular profiling of pediatric mature B-cell lymphoma treated in population-based prospective clinical trials. Blood. 2008;112:1374–81.
Yang Y, Shaffer AL III, Emre NC, Ceribelli M, Zhang M, Wright G, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21:723–37.
Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034.
Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25:275–86.
Frey WD, Chaudhry A, Slepicka PF, Ouellette AM, Kirberger SE, Pomerantz WCK, et al. BPTF maintains chromatin accessibility and the self-renewal capacity of mammary gland stem cells. Stem Cell Rep. 2017;9:23–31.
Xu B, Cai L, Butler JM, Chen D, Lu X, Allison DF, et al. The chromatin remodeler BPTF activates a stemness gene-expression program essential for the maintenance of adult hematopoietic stem cells. Stem Cell Rep. 2018;10:675–83.
Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. Nfkb 1 is dispensable for Myc‐induced lymphomagenesis. Oncogene. 2005;24:6231–40.
Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T. The IKK2/NF-kB pathway suppresses MYC-induced lymphomagenesis. Blood. 2009;114:2448–58.
Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354:2431–42.
Yang Z, Shah K, Busby T, Giles K, Khodadadi-Jamayran A, Li W, et al. Hijacking a key chromatin modulator creates epigenetic vulnerability for MYC-driven cancer. J Clin Investig. 2018;128:3605–18.
Iritani BM, Forbush KA, Farrar MA, Perlmutter RM. Control of B cell development by Ras-mediated activation of Raf. EMBO J. 1997;16:7019–31.
Graña O, Rubio-Camarillo M, Fdez-Riverola F, Pisano DG, Glez-Peña D. Nextpresso: Next generation sequencing expression analysis pipeline. Curr Bioinform. 2018;13:583–91.
Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–99.
Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–59.
Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–49.
Acknowledgements
We thank the CNIO Core facilities and ECG members for support, V. J. Sánchez-Arévalo for valuable discussions, E. Carrillo-de-Santa Pau for discussion on bioinformatics analyses, and V. de Yébenes for critical comments to a previous version of the manuscript. We thank M. Reth and C. Blanco for providing Mb1-Cre and Eμ-Myc mice, respectively. FXR was supported, in part, by grants SAF2011-29530, SAF2015-70553-R, RTI2018-101071-B-I00, and RTI2018-101071-B-I00 (Ministerio de Ciencia, Innovación y Universidades, Madrid, Spain) (co-funded by the ERDF-EU) and RTICC (RD12/0036/0034, RD12/0036/0050, Instituto de Salud Carlos III). AR was supported by ERC StG BCLYM-207844, SAF2013-42767-R, and SAF2016-75511-R grants and co-funding by Fondo Europeo de Desarrollo Regional (FEDER). CNIO is supported by Ministerio de Ciencia, Innovación y Universidades as a Centro de Excelencia Severo Ochoa SEV-2015-0510. The CNIC is supported by the Ministerio de Ciencia, Innovación y Universidades (MCNU) and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505). Personal grants: PD and JP, Asociación Española Contra el Cáncer; IF, Amigos del CNIO/Juegaterapia; MPA, Ministerio de Ciencia, Innovación y Universidades, Spain. FXR acknowledges the support of Asociación Española Contra el Cáncer.
Author information
Authors and Affiliations
Contributions
LR designed and performed the in vivo studies; processed, and analysed the RNA-sequencing data, and analysed the public expression datasets of human aggressive B-cell lymphomas. IF performed the immunohistochemical analysis of tumor samples and the western blot characterization of tumor samples. PD designed, performed, and analysed the BM transplantation experiments. MPA and JP contributed to the conduct of the experimental work. NP provided technical support with mouse colony maintenance. JFG contributed to the histopathological analysis of mouse lymphomas. MAP provided human samples for histological evaluation. AR contributed to experimental design, provided scientific insight, and contributed with reagents and mouse lines. FXR contributed to experimental design, conceived and supervised the study. LR and FXR wrote the paper with contributions from the co-authors. FXR and AR obtained financial support.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Richart, L., Felipe, I., Delgado, P. et al. Bptf determines oncogenic addiction in aggressive B-cell lymphomas. Oncogene 39, 4884–4895 (2020). https://doi.org/10.1038/s41388-020-1331-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-020-1331-3
This article is cited by
-
BPTF-665aa mediate chromatin remodeling drives chemoresistance in T-LBL/ALL
Journal of Experimental & Clinical Cancer Research (2025)
-
BPTF regulates androgen receptor activity by enhancing chromatin accessibility and stabilizing the AR-FOXA1 interaction
Nature Communications (2025)
-
The emerging role of ISWI chromatin remodeling complexes in cancer
Journal of Experimental & Clinical Cancer Research (2021)