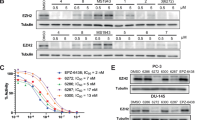
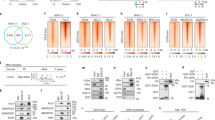
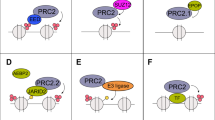
Abstract
Breast cancer (BC) remains the second leading cause of cancer-related mortalities in women. Resistance to hormone therapies such as tamoxifen, an estrogen receptor (ER) inhibitor, is a major hurdle in the treatment of BC. Enhancer of zeste homolog 2 (EZH2), the methyltransferase component of the Polycomb repressive complex 2 (PRC2), has been implicated in tamoxifen resistance. Evidence suggests that EZH2 often functions noncanonically, in a methyltransferase-independent manner, as a transcription coactivator through interacting with oncogenic transcription factors. Unlike methyltransferase inhibitors, proteolysis targeting chimeras (PROTAC) can suppress both activating and repressive functions of EZH2. Here, we find that EZH2 PROTACs, MS177 and MS8815, effectively inhibited the growth of BC cells, including those with acquired tamoxifen resistance, to a much greater degree when compared to methyltransferase inhibitors. Mechanistically, EZH2 associates with forkhead box M1 (FOXM1) and binds to the promoters of FOXM1 target genes. EZH2 PROTACs induce degradation of both EZH2 and FOXM1, leading to reduced expression of target genes involved in cell cycle progression and tamoxifen resistance. Together, this study supports that EZH2-targeted PROTACs represent a promising avenue of research for the future treatment of BC, including in the setting of tamoxifen resistance.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw and processed sequencing files have been submitted to the Gene Expression Omnibus (GEO) database under the accession numbers of GSE270163 and GSE270769.
References
Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49.
Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer. 2004;40:2667–75.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Baranova A, Krasnoselskyi M, Starikov V, Kartashov S, Zhulkevych I, Vlasenko V, et al. Triple-negative breast cancer: current treatment strategies and factors of negative prognosis. J Med Life. 2022;15:153–61.
Kulkarni A, Stroup AM, Paddock LE, Hill SM, Plascak JJ, Llanos AAM. Breast cancer incidence and mortality by molecular subtype: statewide age and racial/ethnic disparities in New Jersey. Cancer Health Disparities. 2019;3:e1–e17.
Yu NY, Iftimi A, Yau C, Tobin NP, van ‘t Veer L, Hoadley KA, et al. Assessment of long-term distant recurrence-free survival associated with tamoxifen therapy in postmenopausal patients with luminal A or luminal B breast cancer. JAMA Oncol. 2019;5:1304–9.
Kuss JT, Muss HB, Hoen H, Case LD. Tamoxifen as initial endocrine therapy for metastatic breast cancer: long term follow-up of two Piedmont Oncology Association (POA) trials. Breast Cancer Res Treat. 1997;42:265–74.
Xu B, Konze KD, Jin J, Wang GG. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp Hematol. 2015;43:698–712.
Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33:903–35.
Guo Y, Zhao S, Wang GG. Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 ‘Readout’, and phase separation-based compaction. Trends Genet. 2021;37:547–65.
Guo Y, Yu Y, Wang GG. Polycomb repressive complex 2 in oncology. Cancer Treat Res. 2023;190:273–320.
Fan T, Jiang S, Chung N, Alikhan A, Ni C, Lee CC, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res. 2011;9:418–29.
Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer cell. 2019;36:385–401 e388.
Wu Y, Zhang Z, Cenciarini ME, Proietti CJ, Amasino M, Hong T, et al. Tamoxifen resistance in breast cancer is regulated by the EZH2-ERalpha-GREB1 transcriptional axis. Cancer Res. 2018;78:671–84.
Wang J, Wang GG. No easy way out for EZH2: its pleiotropic, noncanonical effects on gene regulation and cellular function. Int J Mol Sci. 2020;21:9501.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9.
Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–73.
Bae WK, Yoo KH, Lee JS, Kim Y, Chung IJ, Park MH, et al. The methyltransferase EZH2 is not required for mammary cancer development, although high EZH2 and low H3K27me3 correlate with poor prognosis of ER-positive breast cancers. Mol Carcinog. 2015;54:1172–80.
Bae WK, Hennighausen L. Canonical and non-canonical roles of the histone methyltransferase EZH2 in mammary development and cancer. Mol Cell Endocrinol. 2014;382:593–7.
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT.et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, NY (Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.). 2012;338:1465–9.
Yu X, Wang J, Gong W, Ma A, Shen Y, Zhang C, et al. Dissecting and targeting noncanonical functions of EZH2 in multiple myeloma via an EZH2 degrader. Oncogene. 2023;42:994–1009.
Wang J, Yu X, Gong W, Liu X, Park KS, Ma A, et al. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat Cell Biol. 2022;24:384–99.
Wang J, Park KS, Yu X, Gong W, Earp HS, Wang GG, et al. A cryptic transactivation domain of EZH2 binds AR and AR’s splice variant, promoting oncogene activation and tumorous transformation. Nucleic Acids Res. 2022;50:10929–46.
Hwang C, Giri VN, Wilkinson JC, Wright CW, Wilkinson AS, Cooney KA, et al. EZH2 regulates the transcription of estrogen-responsive genes through association with REA, an estrogen receptor corepressor. Breast Cancer Res Treat. 2008;107:235–42.
Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–19.
Xu H, Xu K, He HH, Zang C, Chen CH, Chen Y, et al. Integrative analysis reveals the transcriptional collaboration between EZH2 and E2F1 in the regulation of cancer-related gene expression. Mol Cancer Res. 2016;14:163–72.
Mahara S, Lee PL, Feng M, Tergaonkar V, Chng WJ, Yu Q. HIFI-alpha activation underlies a functional switch in the paradoxical role of Ezh2/PRC2 in breast cancer. Proc Natl Acad Sci USA. 2016;113:E3735–3744.
Lu XF, Zeng D, Liang WQ, Chen CF, Sun SM, Lin HY. FoxM1 is a promising candidate target in the treatment of breast cancer. Oncotarget. 2018;9:842–52.
Zimmerman SM, Lin PN, Souroullas GP. Non-canonical functions of EZH2 in cancer. Front Oncol. 2023;13:1233953.
Wang C, Chen X, Liu X, Lu D, Li S, Qu L, et al. Discovery of precision targeting EZH2 degraders for triple-negative breast cancer. Eur J Med Chem. 2022;238:114462.
Dale B, Anderson C, Park KS, Kaniskan HU, Ma A, Shen Y, et al. Targeting triple-negative breast cancer by a novel proteolysis targeting chimera degrader of enhancer of zeste homolog 2. ACS Pharm Transl Sci. 2022;5:491–507.
Ma A, Stratikopoulos E, Park KS, Wei J, Martin TC, Yang X, et al. Discovery of a first-in-class EZH2 selective degrader. Nat Chem Biol. 2020;16:214–22.
Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44:6070–86.
Huang L, Zhao S, Frasor JM, Dai Y. An integrated bioinformatics approach identifies elevated cyclin E2 expression and E2F activity as distinct features of tamoxifen resistant breast tumors. PLoS One. 2011;6:e22274.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–76.e516.
Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol cell (Res Support, Non-U S Gov’t). 2011;43:798–810.
Yang L, Kumegawa K, Saeki S, Nakadai T, Maruyama R. Identification of lineage-specific epigenetic regulators FOXA1 and GRHL2 through chromatin accessibility profiling in breast cancer cell lines. Cancer Gene Ther. 2024;31:736–45.
Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic acids Res. 2019;47:W212–W224.
Chen X, Muller GA, Quaas M, Fischer M, Han N, Stutchbury B, et al. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227–36.
Green AR, Aleskandarany MA, Agarwal D, Elsheikh S, Nolan CC, Diez-Rodriguez M, et al. MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br J Cancer. 2016;114:917–28.
Jeffreys SA, Becker TM, Khan S, Soon P, Neubauer H, de Souza P, et al. Prognostic and predictive value of CCND1/Cyclin D1 amplification in breast cancer with a focus on postmenopausal patients: a systematic review and meta-analysis. Front Endocrinol (Lausanne). 2022;13:895729.
Yan J, Li B, Lin B, Lee PT, Chung TH, Tan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood. 2016;128:948–58.
Wan L, Xu K, Wei Y, Zhang J, Han T, Fry C, et al. Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function. Mol Cell. 2018;69:279–91 e275.
Jiao L, Shubbar M, Yang X, Zhang Q, Chen S, Wu Q, et al. A partially disordered region connects gene repression and activation functions of EZH2. Proc Natl Acad Sci USA. 2020;117:16992–7002.
Wang L, Chen C, Song Z, Wang H, Ye M, Wang D, et al. EZH2 depletion potentiates MYC degradation inhibiting neuroblastoma and small cell carcinoma tumor formation. Nat Commun. 2022;13:12.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Lu J, Guo Y, Yin J, Chen J, Wang Y, Wang GG, et al. Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations. Nat Commun. 2024;15:3111.
Li J, Galbo PM Jr., Gong W, Storey AJ, Tsai YH, Yu X, et al. ZMYND11-MBTD1 induces leukemogenesis through hijacking NuA4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism. Nat Commun. 2021;12:1045.
Zhao S, Lu J, Pan B, Fan H, Byrum SD, Xu C, et al. TNRC18 engages H3K9me3 to mediate silencing of endogenous retrotransposons. Nature. 2023;623:633–42.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell. 2018;173:305–20 e310.
Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;2017:PO.17.00073.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
We thank all members of Cai, Jin and Wang laboratories for helpful discussions during the development of this manuscript. We thank Dr. Bryan Pan for bioinformatics support, Dr. Philip Spanheimer for sharing reagents, and John Albright for technical contributions. We thank core facilities of University of North Carolina and Duke University for their professional support in tissue culture, high-throughput sequencing and bioinformatics and data storage.
Funding
This work was supported in part through the UNC Lineberger Comprehensive Cancer Center Core Support Grant P30-CA016086. This work was also supported in part by R01CA262903 (to LC), R01CA218600 (to JJ and GGW), R01CA268519 (to GGW and JJ), R01CA211336 (to GGW) and R01CA230854 (to JJ) grants from the US National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
JJ, LC and GGW designed the experiments and secured fundings. JC and XY led the biological and medicinal chemistry aspects of the study, respectively, under the supervision of GGW, LC and JJ. JC led on the manuscript writing and all authors conducted data analysis and interpretation and participated in the preparation of manuscript.
Corresponding authors
Ethics declarations
Competing interests
JJ is listed as a co-inventor on the patent application (WO 2018/081530 A1) filed by Icahn School of Medicine at Mount Sinai covering EZH2 degraders. JJ is a cofounder and equity shareholder in Cullgen, Inc. and a consultant for Cullgen, Inc., EpiCypher, Inc., and Accent Therapeutics, Inc. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, Inc., Cullgen, Inc., and Cullinan Oncology, Inc.
Ethics declarations
This study does not require ethics approval because no animals and human subjects are used.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Corbin, J., Yu, X., Jin, J. et al. EZH2 PROTACs target EZH2- and FOXM1-associated oncogenic nodes, suppressing breast cancer cell growth. Oncogene 43, 2722–2736 (2024). https://doi.org/10.1038/s41388-024-03119-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03119-9