Abstract
Immunosuppression characterizes the tumour microenvironment in HCC, and recent studies have implicated RNA-binding proteins (RBPs) in the development of HCC. Here, we conducted a screen and identified RBM12 as a key protein that increased the expression of PD-L1, thereby driving immune evasion in HCC. Furthermore, RBM12 was found to be significantly upregulated in HCC tissues and was associated with a poor prognosis for HCC patients. Through various molecular assays and high-throughput screening, we determined that RBM12 could directly bind to the JAK1 mRNA via its 4th-RRM (RNA recognition motif) domain and recruit EIF4A2 through its 2nd-RRM domain, enhancing the distribution of ribosomes on JAK1 mRNA, which promotes the translation of JAK1 and the subsequent upregulation of its expression. As a result, the activated JAK1/STAT1 pathway transcriptionally upregulates PD-L1 expression, facilitating immune evasion in HCC. In summary, our findings provide insights into the significant contribution of RBM12 to immune evasion in HCC, highlighting its potential as a therapeutic target in the future.

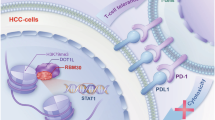
This graphical abstract shows that elevated expression of RBM12 in HCC can augment PD-L1-mediated tumour immune evasion by increasing the efficiency of JAK1 mRNA translation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data associated with this study are presented in the paper or Supplementary Data. Materials supporting the findings of this study are available from the corresponding author upon reasonable request. The high-throughput sequencing data generated in this study are publicly available at SRA datebase (PRJNA1067431).
References
Greten T, Villanueva A, Korangy F, Ruf B, Yarchoan M, Ma L, et al. Biomarkers for immunotherapy of hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2023;20:780–798.
Vitale A, Cabibbo G, Iavarone M, Viganò L, Pinato D, Ponziani F, et al. Personalised management of patients with hepatocellular carcinoma: a multiparametric therapeutic hierarchy concept. Lancet Oncol. 2023;24:e312–e322.
Llovet J, Castet F, Heikenwalder M, Maini M, Mazzaferro V, Pinato D, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–72.
Rimassa L, Finn R, Sangro B. Combination immunotherapy for hepatocellular carcinoma. J Hepatol. 2023;79:506–15.
Sun L, Zhang K, Xie Y, Liu J, Xiao Z. Immunotherapies for advanced hepatocellular carcinoma. Front Pharmacol. 2023;14:1138493.
Personalized DNA Vaccine Tamps Down HCC. Cancer Discov. 2022;12:7–8.
Peiseler M, Schwabe R, Hampe J, Kubes P, Heikenwälder M, Tacke F. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease - novel insights into cellular communication circuits. J Hepatol. 2022;77:1136–60.
Faivre S, Rimassa L, Finn R. Molecular therapies for HCC: Looking outside the box. J Hepatol. 2020;72:342–52.
Cappuyns S, Corbett V, Yarchoan M, Finn R, Llovet J. Critical Appraisal of Guideline Recommendations on Systemic Therapies for Advanced Hepatocellular Carcinoma: A Review. JAMA Oncol. 2023;10:395–404.
Zou Z, Lin Z, Wu C, Tan J, Zhang J, Peng Y, et al. Micro-Engineered Organoid-on-a-Chip Based on Mesenchymal Stromal Cells to Predict Immunotherapy Responses of HCC Patients. Adv Sci (Weinh, Baden-Wurtt, Ger). 2023;10:e2302640.
Pinter M, Scheiner B, Peck-Radosavljevic M. Immunotherapy for advanced hepatocellular carcinoma: a focus on special subgroups. Gut. 2021;70:204–14.
Dhanasekaran R, Nault J, Roberts L, Zucman-Rossi J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology. 2019;156:492–509.
Stark G, Darnell J. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–14.
Danese S, Argollo M, Le Berre C, Peyrin-Biroulet L. JAK selectivity for inflammatory bowel disease treatment: does it clinically matter? Gut. 2019;68:1893–9.
Fountzilas E, Kurzrock R, Vo H, Tsimberidou A. Wedding of Molecular Alterations and Immune Checkpoint Blockade: Genomics as a Matchmaker. J Natl Cancer Inst. 2021;113:1634–47.
Aaronson D, Horvath C. A road map for those who don’t know JAK-STAT. Sci (N. Y, NY). 2002;296:1653–5.
O’Shea J, Gadina M, Schreiber R Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 2002;S121-131.
Cerezo M, Guemiri R, Druillennec S, Girault I, Malka-Mahieu H, Shen S, et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat Med. 2018;24:1877–86.
He S, Valkov E, Cheloufi S, Murn J. The nexus between RNA-binding proteins and their effectors. Nat Rev Genet. 2023;24:276–94.
Wenhui Zhao TL, Ziyi F, Huilin Z, Yangguang DU, Hexu H. NAT10 Promotes the Malignant Progression of Hepatocellular Carcinoma through Upregulating RelA/p65 Acetylation. J Biol Regulators Homeost Agents. 2023;37:2935–46.
Gebauer F, Schwarzl T, Valcárcel J, Hentze M. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22:185–98.
Han H, Lin T, Fang Z, Zhou G. RBM23 Drives Hepatocellular Carcinoma by Activating NF-κB Signaling Pathway. BioMed Res Int. 2021;2021:6697476.
Liu J, Cao X. RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 2023;33:97–115.
Han H, Lin T, Wang Z, Song J, Fang Z, Zhang J, et al. RNA-binding motif 4 promotes angiogenesis in HCC by selectively activating VEGF-A expression. Pharmacol Res. 2023;187:106593.
Kafasla P, Skliris A, Kontoyiannis D. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol. 2014;15:492–502.
Huang Q, Wu X, Wang Z, Chen X, Wang L, Lu Y, et al. The primordial differentiation of tumor-specific memory CD8 T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell. 2022;185:4049–.e4025.
Chen Y, Yang D, Li S, Gao Y, Jiang R, Deng L, et al. Development of a Listeria monocytogenes-based vaccine against hepatocellular carcinoma. Oncogene. 2011;31:2140–52.
Icard P, Simula L, Wu Z, Berzan D, Sogni P, Dohan A, et al. Why may citrate sodium significantly increase the effectiveness of transarterial chemoembolization in hepatocellular carcinoma? Drug Resistance Updates : Rev Commentaries Antimicrobial anticancer Chemother. 2021;59:100790.
Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18:130.
Liu Y, Wu Q, Sun T, Huang J, Han G, Han H. DNAAF5 promotes hepatocellular carcinoma malignant progression by recruiting USP39 to improve PFKL protein stability. Front Oncol. 2022;12:1032579.
Choi W, Yip T, Wong G, Kim W, Yee L, Brooks-Rooney C, et al. Hepatocellular carcinoma risk in patients with chronic hepatitis B receiving tenofovir- vs. entecavir-based regimens: Individual patient data meta-analysis. J Hepatol. 2023;78:534–42.
Yang C, Zhang H, Zhang L, Zhu A, Bernards R, Qin W, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20:203–22.
Topalian S, Forde P, Emens L, Yarchoan M, Smith K, Pardoll D. Neoadjuvant immune checkpoint blockade: A window of opportunity to advance cancer immunotherapy. Cancer Cell. 2023;41:1551–66.
He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660–9.
Topalian S, Drake C, Pardoll D. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61.
Cai L, Li Y, Tan J, Xu L, Li Y. Targeting LAG-3, TIM-3, and TIGIT for cancer immunotherapy. J Hematol Oncol. 2023;16:101.
Pang K, Shi Z, Wei L, Dong Y, Ma Y, Wang W, et al. Research progress of therapeutic effects and drug resistance of immunotherapy based on PD-1/PD-L1 blockade. Drug Resistance Updates : Rev Commentaries Antimicrobial Anticancer Chemother. 2023;66:100907.
Gaikwad S, Agrawal M, Kaushik I, Ramachandran S, Srivastava S. Immune checkpoint proteins: Signaling mechanisms and molecular interactions in cancer immunotherapy. Semin Cancer Biol. 2022;86:137–50.
Xue C, Yao Q, Gu X, Shi Q, Yuan X, Chu Q, et al. Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct Target Ther. 2023;8:204.
Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6:402.
Lin X, Farooqi A. Cucurbitacin mediated regulation of deregulated oncogenic signaling cascades and non-coding RNAs in different cancers: Spotlight on JAK/STAT, Wnt/β-catenin, mTOR, TRAIL-mediated pathways. Semin cancer Biol. 2021;73:302–9.
Endo T, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4.
Pan Y, Shu G, Fu L, Huang K, Zhou X, Gui C, et al. EHBP1L1 Drives Immune Evasion in Renal Cell Carcinoma through Binding and Stabilizing JAK1. Adv Sci (Weinh, Baden-Wurtt, Ger). 2023;10:e2206792.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA mA modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol cell. 2022;82:1660–.e1610.
Han H, Zhu W, Lin T, Liu C, Zhai H. N4BP3 promotes angiogenesis in hepatocellular carcinoma by binding with KAT2B. Cancer Sci. 2022;113:3390–404.
Acknowledgements
We sincerely thank the central Laboratory of Taizhou People’s Hospital Affiliated to Nanjing Medical University for the help of instruments and equipment.
Funding
This project was supported by The Youth Fund of Taizhou People’s Hospital Affiliated to Nanjing Medical University (TZKY20220102), Key research project of Taizhou School of Clinical Medicine, Nanjing Medical University (TZKY20230306), Scientific research start-up fund of Taizhou People’s Hospital (QDJJ202106, QDJJ202103), Taizhou Society Development Project, Jiangsu, China (TS202308), General Project of Jiangsu Provincial Health Commission (H2023030), National Natural Science Foundation of China (82372755, 82203728, 82372746, 82374223), Huzhou Municipal Science and Technology Bureau welfare applied research project (Grant No. 2021GZ69), Research Fund of Anhui Institute of translational medicine (2022zhyx-C21), Outstanding Youth Scientific Research Projects in colleges and universities of Anhui Province (2022AH030115) and Nanjing special foundation for health science and technology development (distinguished young program, JQX21005).
Author information
Authors and Affiliations
Contributions
Qiang Wang, Huimin Guo and Hexu Han performed study concept and design; Hexu Han and Qian Shi completed the experimental part of the project; Yue Zhang, Mingdong Ding and Xianzhong He performed development of methodology and writing, review and revision of the paper; Yin Yuan and Siliang Wang provided acquisition, analysis and interpretation of data, and statistical analysis; Cuixia Liu, Dakun Zhao, Yifan Wang and Yanping Du provided technical and material support. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Three cohorts of HCC tissue samples were collected from the Affiliated Taizhou People’s Hospital of Nanjing Medical University (Jiangsu Province, China), and the experimental protocol was approved by the Institutional Ethics Review Board of the Affiliated Taizhou People’s Hospital of Nanjing Medical University (2022-008-01). Pathological analysis was conducted to validate the authenticity of the specimens. Animal ethical testing certificates were approved by the Experimental Center of Jiangsu Hanjiang Biotechnology Co., LTD (HJSW-23050302), and the animals were raised in accordance with animal welfare laws.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, H., Shi, Q., Zhang, Y. et al. RBM12 drives PD-L1-mediated immune evasion in hepatocellular carcinoma by increasing JAK1 mRNA translation. Oncogene 43, 3062–3077 (2024). https://doi.org/10.1038/s41388-024-03140-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03140-y
This article is cited by
-
RBM8A confers oxaliplatin resistance in gastric cancer by maintaining EGFR mRNA stability
Oncogene (2026)
-
RBM30 recruits DOT1L to activate STAT1 transcription and drive immune evasion in hepatocellular carcinoma
Oncogene (2025)
-
Microglial RUNX1/RBM47 ablation inhibits neuronal ferroptosis via regulating the cGAS-STING-MEF2C pathway in mice with postoperative cognitive dysfunction
Communications Biology (2025)
-
The role of PD‑1/PD‑L1 axis in liver diseases
Clinical and Experimental Medicine (2025)