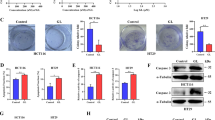
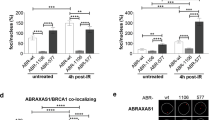
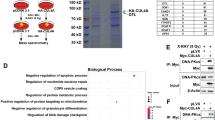
Abstract
Non-homologous end joining (NHEJ), as one major pathway of DNA double-strand break (DSB) repair, could cause genomic instability, which plays pivotal roles in cancer development. While, chromatin remodeling complexes dictate the selection and orchestration of DSB repair pathways by regulating chromatin dynamics. However, the crosstalk between NHEJ and chromatin remodeling in cancer progress remains unclear. In this study, deficiency of GLTSCR1 causes resistance to DNA damage in colorectal cancer (CRC) cells by promoting NHEJ repair efficiency. Mechanistically, GLTSCR1 interacts with BRD9 to engage in the assembly of the non-canonical BAF complex (GBAF). However, GLTSCR1 deficiency disrupts GBAF and triggers the ubiquitination degradation of BRD9. Furthermore, GLTSCR1 deficiency causes aberrant opening in the promoter region of NHEJ repair-associated genes, which promotes CRC development. While, GLTSCR1 and its binding partner BRD9 are not directly involved in assembling NHEJ repair machinery; instead, they regulate the DNA accessibility of NHEJ repair-associated genes. Collectively, our findings confirm GLTSCR1 deficiency as a critical regulatory event of the NHEJ pathway in CRC development, which might require different therapeutic strategy for GLTSCR1 wild-type and mutant CRC.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data generated in this study are available within the article and its supplementary information files. The raw data of the ATAC-seq and RNA-seq data is available in SRA: PRJNA1099778.
References
Centore RC, Sandoval GJ, Soares LMM, Kadoch C, Chan HM. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 2020;36:936–50.
Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304.
Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–22.
Bieluszewski T, Prakash S, Roule T, Wagner D. The role and activity of SWI/SNF chromatin remodelers. Annu Rev Plant Biol. 2023;74:139–63.
Zaret KS. Pioneer transcription factors initiating gene network changes. Annu Rev Genet. 2020;54:367–85.
Tsai S, Fournier LA, Chang EY, Wells JP, Minaker SW, Zhu YD, et al. ARID1A regulates R-loop associated DNA replication stress. PLoS Genet. 2021;17:e1009238.
Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447.
Gatchalian J, Malik S, Ho J, Lee DS, Kelso TWR, Shokhirev MN, et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat Commun. 2018;9:5139.
Alpsoy A, Dykhuizen EC. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem. 2018;293:3892–903.
Helming KC, Wang X, Roberts CWM. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–17.
Han F, Zhang L, Chen C, Wang Y, Zhang Y, Qian L, et al. GLTSCR1 negatively regulates BRD4-dependent transcription elongation and inhibits CRC metastasis. Adv Sci (Weinh). 2019;6:1901114.
Han F, Yang B, Zhou M, Huang Q, Mai M, Huang Z, et al. GLTSCR1 coordinates alternative splicing and transcription elongation of ZO1 to regulate colorectal cancer progression. J Mol Cell Biol. 2022;14:mjac009.
Perkhofer L, Gout J, Roger E, Kude de Almeida F, Baptista Simoes C, Wiesmuller L, et al. DNA damage repair as a target in pancreatic cancer: state-of-the-art and future perspectives. Gut. 2021;70:606–17.
Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8.
Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6:166–75.
Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714.
Stinson BM, Loparo JJ. Repair of DNA double-strand breaks by the nonhomologous end joining pathway. Annu Rev Biochem. 2021;90:137–64.
Tang M, Chen G, Tu B, Hu Z, Huang Y, DuFort CC, et al. SMYD2 inhibition-mediated hypomethylation of Ku70 contributes to impaired nonhomologous end joining repair and antitumor immunity. Sci Adv. 2023;9:eade6624.
Luo J, Chen JW, Zhou J, Han K, Li S, Duan JL, et al. TBX20 inhibits colorectal cancer tumorigenesis by impairing NHEJ-mediated DNA repair. Cancer Sci. 2022;113:2008–21.
Patterson-Fortin J, Bose A, Tsai WC, Grochala C, Nguyen H, Zhou J, et al. Targeting DNA repair with combined inhibition of NHEJ and MMEJ induces synthetic lethality in TP53-mutant cancers. Cancer Res. 2022;82:3815–29.
Bilbao C, Ramirez R, Rodriguez G, Falcon O, Leon L, Diaz-Chico N, et al. Double strand break repair components are frequent targets of microsatellite instability in endometrial cancer. Eur J Cancer. 2010;46:2821–7.
Li ZW, Zhao JM, Tang Y. Advances in the role of SWI/SNF complexes in tumours. J Cell Mol Med. 2023;27:1023–31.
Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 2017;357:1294–8.
Vazquez-Arreguin K, Bensard C, Schell JC, Swanson E, Chen X, Rutter J, et al. Oct1/Pou2f1 is selectively required for colon regeneration and regulates colon malignancy. PLoS Genet. 2019;15:e1007687.
Mashtalir N, Suzuki H, Farrell DP, Sankar A, Luo J, Filipovski M, et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell. 2020;183:802–17.e24.
Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife. 2017;6:e30506.
Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017;549:548–52.
Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125–36.e3.
Lin SH, Raju GS, Huff C, Ye Y, Gu J, Chen JS, et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut. 2018;67:1299–305.
Michel BC, D’Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410.
Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202.
Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15.
Mashtalir N, Dao HT, Sankar A, Liu H, Corin AJ, Bagert JD, et al. Chromatin landscape signals differentially dictate the activities of mSWI/SNF family complexes. Science. 2021;373:306–15.
Rim EY, Clevers H, Nusse R. The Wnt pathway: from signaling mechanisms to synthetic modulators. Annu Rev Biochem. 2022;91:571–98.
Vasileiou G, Ekici AB, Uebe S, Zweier C, Hoyer J, Engels H, et al. Chromatin-remodeling-factor ARID1B represses Wnt/beta-catenin signaling. Am J Hum Genet. 2015;97:445–56.
Wischhof L, Lee HM, Tutas J, Overkott C, Tedt E, Stork M, et al. BCL7A-containing SWI/SNF/BAF complexes modulate mitochondrial bioenergetics during neural progenitor differentiation. EMBO J. 2022;41:e110595.
Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–4.
Haber JE. A life investigating pathways that repair broken chromosomes. Annu Rev Genet. 2016;50:1–28.
Makharashvili N, Paull TT. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair. 2015;32:75–81.
Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–20.
Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol. 2018;15:564–76.
Wu JN, Roberts CWM. Mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43.
Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;74:2465–75.
Zhou Q, Huang J, Zhang C, Zhao F, Kim W, Tu X, et al. The bromodomain containing protein BRD-9 orchestrates RAD51-RAD54 complex formation and regulates homologous recombination-mediated repair. Nat Commun. 2020;11:2639.
Acknowledgements
We thank Qiong Huang from the core facility platform of Zhejiang University School of Medicine for technical support. We thank the Laboratory Animal Center of Zhejiang University for technical assistance with management of mice.
Funding
This work was supported by National Natural Science Foundation of China (grants 81871937, 82472952 and 82173223), the CAMS Innovation Fund for Medical Sciences (CIFMS, grant number 2019-I2M-5-044) and the Guangdong Basic and Applied Basic Research Foundation (grant number 2023A1515140182).
Author information
Authors and Affiliations
Contributions
Conceptualization, HZ, FH, EX and ZT; methodology, FH, XZ, and LL; investigation, BY, PL and LL; writing—original draft, FH, XZ; writing—review & editing, HZ, FH, EX and ZT; funding acquisition, HZ and FH; resources, HZ and FH; Supervision, HZ and FH and all authors have approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods in this study were performed in accordance with the Declaration of Helsinki and its subsequent revisions. Colorectal carcinoma samples from patients in this study were collected from the Run Run Shaw Hospital, Zhejiang University. All participating patients were informed. The study was approved by the Ethic Committee of Zhejiang University, school of Medicine (Approval number: 2018‐018) and informed consent was obtained from every participant.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, F., Zhou, X., Liu, L. et al. GLTSCR1 deficiency promotes colorectal cancer development through regulating non-homologous end joining. Oncogene 43, 3517–3531 (2024). https://doi.org/10.1038/s41388-024-03179-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03179-x