Abstract
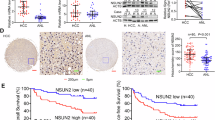
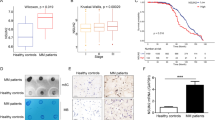
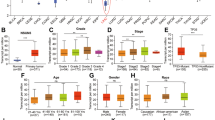
5-Methylcytosine (m5C) RNA modification is a highly abundant and important epigenetic modification in mammals. As an important RNA m5C methyltransferase, NOP2/Sun-domain family member 2 (NSUN2)-mediated m5C RNA modification plays an important role in the regulation of the biological functions in many cancers. However, little is known about the biological role of NSUN2 in hepatocellular carcinoma (HCC). In this study, we found that the expression of NSUN2 was significantly upregulated in HCC, and the HCC patients with higher expression of NSUN2 had a poorer prognosis than those with lower expression of NSUN2. NSUN2 could affect the tumor immune regulation of HCC in several ways. In vitro and in vivo experiments confirmed that NSUN2 knockdown significantly decreased the abilities of proliferation, colony formation, migration and invasion of HCC cells. The methylated RNA immunoprecipitation-sequencing (MeRIP-seq) showed NSUN2 knockdown significantly affected the abundance, distribution, and composition of m5C RNA modification in HCC cells. Functional enrichment analyses and in vitro experiments suggested that NSUN2 could promote the HCC cells to proliferate, migrate and invade by regulating Wnt signaling pathway. SARS2 were identified via the RNA immunoprecipitation-sequencing (RIP-Seq) and MeRIP-seq as downstream target of NSUN2, which may play an important role in tumor-promoting effect of NSUN2-mediated m5C RNA modification in HCC. In conclusion, NSUN2 promotes HCC progression by regulating Wnt signaling pathway and SARS2 in an m5C-dependent manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data related to this study can be obtained from the corresponding authors upon reasonable request. The m5C MeRIP-seq data are available in GEO database (ID: GSE228973), the single cell transcriptome sequencing data are available in GSE149614 from GEO database, and other RNA-seq data are available in TCGA database (TCGA-LIHC).
References
Freddie B, Mathieu L, Hyuna S, Jacques F, Rebecca LS, Isabelle S, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63.
Amit GS, Masatoshi K, Jordi B. Breakthroughs in hepatocellular carcinoma therapies. Clin Gastroenterol Hepatol. 2023;21:2135–49.
Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2019;17:139–52.
Josep ML, Robin Kate K, Augusto V, Amit GS, Eli P, Sasan R, et al. Hepatocellular carcinoma. Nat Rev Dis Prim. 2021;7:6.
Lehrich BM, Zhang J, Monga SP, Dhanasekaran R. Battle of the biopsies: role of tissue and liquid biopsy in hepatocellular carcinoma. J Hepatol. 2023;80:515–30.
Edward AM, Caroline RD, Feng-Chun Y. ASXL1/2 mutations and myeloid malignancies. J Hematol Oncol. 2022;15:127.
Xiaoxuan W, Yuheng H, Shen M, Wenchen G, Tianyuan R, Tingting Z, et al. A novel immune-related epigenetic signature based on the transcriptome for predicting the prognosis and therapeutic response of patients with diffuse large B-cell lymphoma. Clin Immunol. 2022;243:109105.
Yu Y, Diguang W, Lu Z, Jiao L, Xiao X, Yucheng C, et al. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. 2022;18:5001–18.
Weifeng Y, Xue H, Mian G, Chaojin C, Xue X, Haobo L, et al. N(6)-methyladenosine (m(6)A) methylation in ischemia-reperfusion injury. Cell Death Dis. 2020;11:478.
Huanxiang C, Hongyang L, Chenxing Z, Nan X, Yang L, Xiangzhuan Z, et al. RNA methylation-related inhibitors: Biological basis and therapeutic potential for cancer therapy. Clin Transl Med. 2024;14:e1644.
Chen X, Qingfei C, Qiuxian Z, Shiman J, Zhengyi B, Yuanshuai S, et al. Role of main RNA modifications in cancer: N(6)-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct Target Ther. 2022;7:142.
Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2016;18:31–42.
Guo G, Pan K, Fang S, Ye L, Tong X, Wang Z, et al. Advances in mRNA 5-methylcytosine modifications: detection, effectors, biological functions, and clinical relevance. Mol Ther Nucleic Acids. 2021;26:575–93.
Chen Y, Jiang Z, Yang Y, Zhang C, Liu H, Wan J. The functions and mechanisms of post-translational modification in protein regulators of RNA methylation: current status and future perspectives. Int J Biol Macromol. 2023;253:126773.
Cheng M, You X, Ding J, Dai Y, Chen M, Yuan B, et al. Novel dual methylation of cytidines in the RNA of mammals. Chem Sci. 2021;12:8149–56.
Pan J, Huang Z, Xu Y. m5C-Related lncRNAs predict overall survival of patients and regulate the tumor immune microenvironment in lung adenocarcinoma. Front Cell Dev Biol. 2021;9:671821.
Zou S, Huang Y, Yang Z, Zhang J, Meng M, Zhang Y, et al. NSUN2 promotes colorectal cancer progression by enhancing SKIL mRNA stabilization. Clin Transl Med. 2024;14:e1621.
Zhang Y, Chen X, Zhang H, Wen J, Gao H, Shi B, et al. CDK13 promotes lipid deposition and prostate cancer progression by stimulating NSUN5-mediated m5C modification of ACC1 mRNA. Cell Death Differ. 2023;30:2462–76.
Zhu W, Wan F, Xu W, Liu Z, Wang J, Zhang H, et al. Positive epigenetic regulation loop between AR and NSUN2 promotes prostate cancer progression. Clin Transl Med. 2022;12:e1028.
Chen S, Chen K, Ding L, Yu C, Wu H, Chou Y, et al. RNA bisulfite sequencing reveals NSUN2-mediated suppression of epithelial differentiation in pancreatic cancer. Oncogene. 2022;41:3162–76.
Wu R, Sun C, Chen X, Yang R, Luan Y, Zhao X, et al. NSUN5/TET2-directed chromatin-associated RNA modification of 5-methylcytosine to 5-hydroxymethylcytosine governs glioma immune evasion. Proc Natl Acad Sci USA. 2024;121:e2321611121.
Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin Y, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019;110:3510–9.
Zhen S, Shonglei X, Hui X, Xuming H, Shihao C, Zhe Y, et al. Expression profiles of long noncoding RNAs associated with the NSUN2 gene in HepG2 cells. Mol Med Rep. 2019;19:2999–3008.
Min Y, Renxiong W, Sheng Z, Sang H, Xiaoxiao L, Zhiqiang Y, et al. NSUN2 promotes osteosarcoma progression by enhancing the stability of FABP5 mRNA via m(5)C methylation. Cell Death Dis. 2023;14:125.
Ning W, Ri-Xin C, Min-Hua D, Wen-Su W, Zhao-Hui Z, Kang N, et al. m(5)C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis. 2023;14:139.
Yueqin W, Jingyao W, Luyao F, Ouwen L, Lan H, Shaoxuan Z, et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. 2023;22:81.
Ying L, Yiwen X, Tianlu J, Zetian C, Yikai S, Jie L, et al. Long noncoding RNA DIAPH2-AS1 promotes neural invasion of gastric cancer via stabilizing NSUN2 to enhance the m5C modification of NTN1. Cell Death Dis. 2023;14:260.
Shuai-Jun C, Jun Z, Ting Z, Shan-Shan R, Qian L, Ling-Yan X, et al. Epigenetically upregulated NSUN2 confers ferroptosis resistance in endometrial cancer via m(5)C modification of SLC7A11 mRNA. Redox Biol. 2023;69:102975.
Peng L, Wenlong W, Ruixin Z, Ying D, Xinying L. The m(5) C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin Transl Med. 2023;13:e1466.
Xuhui T, Rong T, Mingming X, Jin X, Wei W, Bo Z, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174.
Anna G-M, Malgorzata C. WNT/β-catenin signaling in hepatocellular carcinoma: The aberrant activation, pathogenic roles, and therapeutic opportunities. Genes Dis. 2023;11:727–46.
Chen X, Qinfan Y, Xinyu G, Qingmiao S, Xin Y, Qingfei C, et al. Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct Target Ther. 2023;8:204.
Zou G, Park J. Wnt signaling in liver regeneration, disease, and cancer. Clin Mol Hepatol. 2023;29:33–50.
Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99.
Shah K, Panchal S, Patel B. Porcupine inhibitors: Novel and emerging anti-cancer therapeutics targeting the Wnt signaling pathway. Pharmacol Res. 2021;167:105532.
Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2015;44:852–62.
Shobbir H. The emerging roles of Cytosine-5 methylation in mRNAs. Trends Genet. 2021;37:498–500.
Saori N, Takeo S, Layla K, Hiroyoshi I, Kana A, Tsutomu S. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546–51.
Mayumi O, Mamoru F, Masato H, Kaoru O, Futoshi Y, Hiroaki K, et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014;10:e1004639.
Jianheng L, Tao H, Yusen Z, Tianxuan Z, Xueni Z, Wanying C, et al. Sequence- and structure-selective mRNA m(5)C methylation by NSUN6 in animals. Natl Sci Rev. 2021;8:nwaa273.
Xiaotian Z, Zhenyun L, Jie Y, Hao T, Junyue X, Minqwei Y, et al. The tRNA methyltransferase NSun2 stabilizes p16INK4 mRNA by methylating the 3’-untranslated region of p16. Nat Commun. 2012;3:712.
Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842.
Chen L, Ding J, Wang B, Chen X, Ying X, Yu Z, et al. RNA methyltransferase NSUN2 promotes hypopharyngeal squamous cell carcinoma proliferation and migration by enhancing TEAD1 expression in an mC-dependent manner. Exp Cell Res. 2021;404:112664.
Chen S, Zhang J, Zhou T, Rao S, Li Q, Xiao L, et al. Epigenetically upregulated NSUN2 confers ferroptosis resistance in endometrial cancer via mC modification of SLC7A11 mRNA. Redox Biol. 2024;69:102975.
Liangliang X, Chang Z, Shanshan Z, Timothy Shun Man C, Yan Z, Weiwei C, et al. Reshaping the systemic tumor immune environment (STIE) and tumor immune microenvironment (TIME) to enhance immunotherapy efficacy in solid tumors. J Hematol Oncol. 2022;15:87.
Benjamin R, Matthias B, Sepideh B, Noemi K, Lichun M, Mahler R, et al. Tumor-associated macrophages trigger MAIT cell dysfunction at the HCC invasive margin. Cell. 2023;186:3686–3705.e3632.
Mikhail B, Edward WR, Kelly K, Vincent C, Douglas FF, Miriam M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50.
de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403.
Qingqing Q, Ying Z, Jintao G, Qinwei C, Weiwei T, Yuchen L, et al. Conserved methylation signatures associate with the tumor immune microenvironment and immunotherapy response. Genome Med. 2024;16:47.
Feng-Ming T, Hsuan-Hsuan L, Shu-Yung L, Hsing-Chen T. Epigenetic remodeling of the immune landscape in cancer: therapeutic hurdles and opportunities. J Biomed Sci. 2023;30:3.
Zaoqu L, Haijiao Z, Qin D, Hui X, Long L, Yuyuan Z, et al. Biological and pharmacological roles of m(6)A modifications in cancer drug resistance. Mol Cancer. 2022;21:220.
Ganglei L, Qinfan Y, Peixi L, Hongfei Z, Yingjun L, Sichen L, et al. Critical roles and clinical perspectives of RNA methylation in cancer. MedComm (2020). 2024;5:e559.
Magen A, Hamon P, Fiaschi N, Soong B, Park M, Mattiuz R, et al. Intratumoral dendritic cell-CD4 T helper cell niches enable CD8 T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat Med. 2023;29:1389–99.
Lai I, Swaminathan S, Baylot V, Mosley A, Dhanasekaran R, Gabay M, et al. Lipid nanoparticles that deliver IL-12 messenger RNA suppress tumorigenesis in MYC oncogene-driven hepatocellular carcinoma. J Immunother cancer. 2018;6:125.
Aghayev T, Mazitova A, Fang J, Peshkova I, Rausch M, Hung M, et al. IL27 signaling serves as an immunologic checkpoint for innate cytotoxic cells to promote hepatocellular carcinoma. Cancer Discov. 2022;12:1960–83.
Huang R, Wang Z, Hong J, Wu J, Huang O, He J, et al. Targeting cancer-associated adipocyte-derived CXCL8 inhibits triple-negative breast cancer progression and enhances the efficacy of anti-PD-1 immunotherapy. Cell Death Dis. 2023;14:703.
Feng D, Xiaoyang Q, Baofeng W, Qian L, Jinyang H, Xuan C, et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021;81:5876–88.
Hosein A, Dangol G, Okumura T, Roszik J, Rajapakshe K, Siemann M, et al. Loss of Rnf43 Accelerates Kras-Mediated Neoplasia and Remodels the Tumor Immune Microenvironment in Pancreatic Adenocarcinoma. Gastroenterology. 2022;162:1303–18.e1318.
Yuqiu X, Zhuang W, Mei F, Dexiang Z, Shenglin M, Zhongen W, et al. Tumor-infiltrated activated B cells suppress liver metastasis of colorectal cancers. Cell Rep. 2022;40:111295.
Biasci D, Smoragiewicz M, Connell CM, Wang Z, Gao Y, Thaventhiran JED, et al. CXCR4 inhibition in human pancreatic and colorectal cancers induces an integrated immune response. Proc Natl Acad Sci USA. 2020;117:28960–70.
Peng DH, Rodriguez BL, Diao L, Chen L, Wang J, Byers LA, et al. Collagen promotes anti-PD-1/PD-L1 resistance in cancer through LAIR1-dependent CD8(+) T cell exhaustion. Nat Commun. 2020;11:4520.
Qiuyao W, Pu T, Dasa H, Zhenchang J, Yunfei H, Wenqian L, et al. SCUBE2 mediates bone metastasis of luminal breast cancer by modulating immune-suppressive osteoblastic niches. Cell Res. 2023;33:464–78.
Horn LA, Chariou PL, Gameiro SR, Qin H, Iida M, Fousek K, et al. Remodeling the tumor microenvironment via blockade of LAIR-1 and TGF-β signaling enables PD-L1-mediated tumor eradication. J Clin Investig. 2022;132:e155148.
Qingyu D, Shunhao Z, Haotian Z, Jing S, Jing L, Guihua W, et al. MARCO is a potential prognostic and immunotherapy biomarker. Int Immunopharmacol. 2023;116:109783.
Thapa B, Kato S, Nishizaki D, Miyashita H, Lee S, Nesline MK, et al. OX40/OX40 ligand and its role in precision immune oncology. Cancer Metastasis Rev. 2024;43:1001–13.
Charlotte L, Maud K, Sophie V, Walid C, Anne S, Claire M, et al. Immune gene expression in head and neck squamous cell carcinoma patients. Eur J Cancer. 2019;121:210–23.
Jiaqi L, Qing X, Jiani X, Chenxi N, Yuanyuan L, Xiaojun Z, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7:3.
Wenqiang Z, Kexin Z, Yanhui M, Yixin S, Tongbing Q, Guoji X, et al. Secreted frizzled-related proteins: A promising therapeutic target for cancer therapy through Wnt signaling inhibition. Biomed Pharmacother. 2023;166:115344.
Yu M, Qin K, Fan J, Zhao G, Zhao P, Zeng W, et al. The evolving roles of Wnt signaling in stem cell proliferation and differentiation, the development of human diseases, and therapeutic opportunities. Genes Dis. 2024;11:101026.
Perugorria M, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin J, et al. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16:121–36.
Xiang D, Gu M, Liu J, Dong W, Yang Z, Wang K, et al. m6A RNA methylation-mediated upregulation of HLF promotes intrahepatic cholangiocarcinoma progression by regulating the FZD4/β-catenin signaling pathway. Cancer Lett. 2023;560:216144.
Mohamad HS, Wenli S. Survey on multi-omics, and multi-omics data analysis, integration and application. Curr Pharm Anal. 2023;19:267–81.
Lu Y, Yang A, Quan C, Pan Y, Zhang H, Li Y, et al. A single-cell atlas of the multicellular ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun. 2022;13:4594.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Chen B, Khodadoust M, Liu C, Newman A, Alizadeh A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59.
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12:21.
Anton S, Tingting J, Giampaolo B, Balázs G, Thomas K, Christos H, et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Res. 2017;77:3317–24.
Beibei R, Ching Ngar W, Yin T, Jia Yi Z, Sophia Shek Wa Z, Wai Chung W, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2.
Fromowitz FB, Viola MV, Chao S, Oravez S, Mishriki Y, Finkel G, et al. ras p21 expression in the progression of breast cancer. Hum Pathol. 1987;18:1268–75.
Lichinchi G, Gao S, Saletore Y, Gonzalez G, Bansal V, Wang Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1:16011.
Andrey K, Uljana B, Alexander K, Maxim F. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24:1138–43.
Daehwan K, Joseph MP, Chanhee P, Christopher B, Steven LS. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15.
Heng L, Bob H, Alec W, Tim F, Jue R, Nils H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Li S, Ning-Yi S, Xiaochuan L, Ian M, Jian F, Eric JN. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8:e65598.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9.
Chi S, Zang J, Mele A, Darnell R. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86.
Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Acknowledgements
This work was supported by the China Postdoctoral Science Foundation (2023M743200 and 2024T170826), the Postdoctoral Fellowship Program of CPSF (GZC20232425), the Science and Technology Research Program of Henan Province (242102311163), the Henan Medical Science and Technology Joint Building Program (LHGJ20230160), and the Henan Sunshine Medical Health Development Foundation iGanDan Project (HKP2023001 and HKP2023006).
Author information
Authors and Affiliations
Contributions
CX and YH conceptualized and supervised this study, HX and XG participated in the overall experiments and wrote the draft. YL prepared figures. LX collected and analyzed data. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods used in this study were performed in accordance with the relevant guidelines and regulations. All animal experiments in this study were carried out in accordance with the regulations of Experimental Animal Use and Welfare Committee of Zhengzhou University (approval number: ZZU-LAC2024060416).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xing, H., Gu, X., Liu, Y. et al. NSUN2 regulates Wnt signaling pathway depending on the m5C RNA modification to promote the progression of hepatocellular carcinoma. Oncogene 43, 3469–3482 (2024). https://doi.org/10.1038/s41388-024-03184-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03184-0
This article is cited by
-
METTL5 promotes tumor progression and ferroptosis resistance via MGST1 in HCC
Molecular and Cellular Biochemistry (2026)
-
LINC02167 stabilizes KSR1 mRNA in an m5C-dependent manner to regulate the ERK/MAPK signaling pathway and promotes colorectal cancer metastasis
Journal of Experimental & Clinical Cancer Research (2025)
-
NSUN2 knockdown ameliorates hepatic glucose and lipid metabolism disorders in type 2 diabetes mellitus through the Inhibition of ACSL6 m5C methylation
Lipids in Health and Disease (2025)
-
NSUN2–tRNAVal−CAC-axis-regulated codon-biased translation drives triple-negative breast cancer glycolysis and progression
Cellular & Molecular Biology Letters (2025)
-
m5C RNA methylation in cancer: from biological mechanism to clinical perspectives
European Journal of Medical Research (2025)