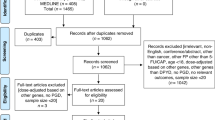
Abstract
Testing for four dihydropyrimidine dehydrogenase (DPYD) variants (DPYD*2 A, DPYD*13, c.2846 A > T, DPYD-HapB3) is currently implemented in clinical practice to prevent fluoropyrimidines (FLs) related toxicity but with limited sensitivity. This study aimed to identify novel genetic factors in FL-related genes to enhance risk prediction using data from the PREPARE trial (NCT03093818). Two hundred seventy-four patients receiving FL-based chemotherapy with severe toxicity were sequenced for 60 candidate genes. Gene and pathway-level association analyses focusing mainly on rare variants were performed using dedicated statistical tests, including gene-wise variant burden (GVB) analysis. DPYD germline variant burden beyond the four routinely tested markers emerged to contribute to toxicity, indicating that rarer genetic variants could help in refining the optimal FL dosage (p < 0.1). Functional rare variant burden in ABCB5, PARP1, ENOSF1, CYP3A4 and nuclear receptors pathway impacted on toxicity risk (p < 0.05 in at least one statistical test). GVB analysis confirmed ABCB5 as a significant risk gene and highlighted ABCC4, HNF4A, and XRCC3 as additional candidates. A predictive model combining genetic burden scores with clinical variables improved the identification of high-risk patients (sensitivity=0.71, specificity=0.74, accuracy=0.73). This study indicated a paradigm shift from population to individual-level arguing for an extension of testing beyond the four DPYD currently considered variants to predict FL-related toxicity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
De Mattia E, Milan N, Assaraf YG, Toffoli G, Cecchin E. Clinical Implementation of Rare and Novel DPYD Variants for Personalizing Fluoropyrimidine Treatment: Challenges and Opportunities. Int J Biol Sci. 2024;20:3742–59.
Lauschke VM, Zhou Y, Ingelman-Sundberg M. Pharmacogenomics Beyond Single Common Genetic Variants: The Way Forward. Annu Rev Pharm Toxicol. 2024;64:33–51.
Peruzzi E, Roncato R, De Mattia E, Bignucolo A, Swen JJ, Guchelaar HJ, et al. Implementation of pre-emptive testing of a pharmacogenomic panel in clinical practice: Where do we stand?. Br J Clin Pharm. 2025;91:270–82.
Zhou Y, Tremmel R, Schaeffeler E, Schwab M, Lauschke VM. Challenges and opportunities associated with rare-variant pharmacogenomics. Trends Pharm Sci. 2022;43:852–65.
Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2017;19:20–9.
Zhou Y, Dagli Hernandez C, Lauschke VM. Population-scale predictions of DPD and TPMT phenotypes using a quantitative pharmacogene-specific ensemble classifier. Br J Cancer. 2020;123:1782–9.
Knikman JE, Gelderblom H, Beijnen JH, Cats A, Guchelaar HJ, Henricks LM. Individualized Dosing of Fluoropyrimidine-Based Chemotherapy to Prevent Severe Fluoropyrimidine-Related Toxicity: What Are the Options?. Clin Pharm Ther. 2021;109:591–604.
Lunenburg C, Henricks LM, Guchelaar HJ, Swen JJ, Deenen MJ, Schellens JHM, et al. Prospective DPYD genotyping to reduce the risk of fluoropyrimidine-induced severe toxicity: Ready for prime time. Eur J Cancer. 2016;54:40–8.
Dalle Fratte C, Polesel J, Roncato R, De Mattia E, Ecca F, Bignucolo A, et al. DPYD Gene Activity Score Predicts Dose-Limiting Toxicity in Fluoropyrimidine-Treated Colorectal Cancer Patients. J Mol Clin Med. 2018;1:143–9.
Fragoulakis V, Roncato R, Bignucolo A, Patrinos GP, Toffoli G, Cecchin E, et al. Cost-utility analysis and cross-country comparison of pharmacogenomics-guided treatment in colorectal cancer patients participating in the U-PGx PREPARE study. Pharm Res. 2023;197:106949.
Fragoulakis V, Roncato R, Fratte CD, Ecca F, Bartsakoulia M, Innocenti F, et al. Estimating the Effectiveness of DPYD Genotyping in Italian Individuals Suffering from Cancer Based on the Cost of Chemotherapy-Induced Toxicity. Am J Hum Genet. 2019;104:1158–68.
Roncato R, Bignucolo A, Peruzzi E, Montico M, De Mattia E, Foltran L, et al. Clinical Benefits and Utility of Pretherapeutic DPYD and UGT1A1 Testing in Gastrointestinal Cancer: A Secondary Analysis of the PREPARE Randomized Clinical Trial. JAMA Netw Open. 2024;7:e2449441.
Toffoli G, Giodini L, Buonadonna A, Berretta M, De Paoli A, Scalone S, et al. Clinical validity of a DPYD-based pharmacogenetic test to predict severe toxicity to fluoropyrimidines. Int J Cancer. 2015;137:2971–80.
Toffoli G, Innocenti F, Polesel J, De Mattia E, Sartor F, Dalle Fratte C, et al. The Genotype for DPYD Risk Variants in Patients With Colorectal Cancer and the Related Toxicity Management Costs in Clinical Practice. Clin Pharm Ther. 2019;105:994–1002.
Amstutz U, Henricks LM, Offer SM, Barbarino J, Schellens JHM, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharm Ther. 2018;103:210–6.
Bignucolo A, De Mattia E, Roncato R, Peruzzi E, Scarabel L, D’Andrea M, et al. Ten-year experience with pharmacogenetic testing for DPYD in a national cancer center in Italy: Lessons learned on the path to implementation. Front Pharm. 2023;14:1199462.
de With M, Sadlon A, Cecchin E, Haufroid V, Thomas F, Joerger M, et al. Implementation of dihydropyrimidine dehydrogenase deficiency testing in Europe. ESMO Open. 2023;8:101197.
Lunenburg C, van der Wouden CH, Nijenhuis M, Crommentuijn-van Rhenen MH, de Boer-Veger NJ, Buunk AM, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur J Hum Genet. 2020;28:508–17.
Lopes JL, Harris K, Karow MB, Peterson SE, Kluge ML, Kotzer KE, et al. Targeted Genotyping in Clinical Pharmacogenomics: What Is Missing?. J Mol Diagn. 2022;24:253–61.
Larrue R, Fellah S, Hennart B, Sabaouni N, Boukrout N, Van der Hauwaert C, et al. Integrating rare genetic variants into DPYD pharmacogenetic testing may help preventing fluoropyrimidine-induced toxicity. Pharmacogenomics J. 2024;24:1.
Knikman JE, Zhai Q, Lunenburg C, Henricks LM, Bohringer S, van der Lee M, et al. Discovering novel germline genetic variants linked to severe fluoropyrimidine-related toxicity in- and outside DPYD. Genome Med. 2024;16:101.
De Mattia E, Silvestri M, Polesel J, Ecca F, Mezzalira S, Scarabel L, et al. Rare genetic variant burden in DPYD predicts severe fluoropyrimidine-related toxicity risk. Biomed Pharmacother. 2022;154:113644.
De Mattia E, Polesel J, Silvestri M, Roncato R, Scarabel L, Calza S, et al. The burden of rare variants in DPYS gene is a novel predictor of the risk of developing severe fluoropyrimidine-related toxicity. Hum Genom. 2023;17:99.
Swen JJ, van der Wouden CH, Manson LE, Abdullah-Koolmees H, Blagec K, Blagus T, et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet. 2023;401:347–56.
van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Davila-Fajardo CL, Deneer VH, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharm Ther. 2017;101:341–58.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997v2 [q-bioGN]. 2013.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Zhou Y, Mkrtchian S, Kumondai M, Hiratsuka M, Lauschke VM. An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 2019;19:115–26.
Zhou Y, Pirmann S, Lauschke VM. APF2: an improved ensemble method for pharmacogenomic variant effect prediction. Pharmacogenomics J. 2024;24:17.
Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23.
Park Y, Kim H, Choi JY, Yun S, Min BJ, Seo ME, et al. Star Allele-Based Haplotyping versus Gene-Wise Variant Burden Scoring for Predicting 6-Mercaptopurine Intolerance in Pediatric Acute Lymphoblastic Leukemia Patients. Front Pharm. 2019;10:654.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Gaedigk A, Casey ST, Whirl-Carrillo M, Miller NA, Klein TE. Pharmacogene Variation Consortium: A Global Resource and Repository for Pharmacogene Variation. Clin Pharm Ther. 2021;110:542–5.
Meulendijks D, Henricks LM, Sonke GS, Deenen MJ, Froehlich TK, Amstutz U, et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: a systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015;16:1639–50.
Bukhari N, Azam F, Alfawaz M, Zahrani M. Identifying a Novel DPYD Polymorphism Associated with Severe Toxicity to 5-FU Chemotherapy in a Saudi Patient. Case Rep Genet. 2019;2019:5150725.
Del Re M, Michelucci A, Di Leo A, Cantore M, Bordonaro R, Simi P, et al. Discovery of novel mutations in the dihydropyrimidine dehydrogenase gene associated with toxicity of fluoropyrimidines and viewpoint on preemptive pharmacogenetic screening in patients. EPMA J. 2015;6:17.
Garcia-Gonzalez X, Lopez-Tarruella S, Garcia MI, Gonzalez-Haba E, Blanco C, Salvador-Martin S, et al. Severe toxicity to capecitabine due to a new variant at a donor splicing site in the dihydropyrimidine dehydrogenase (DPYD) gene. Cancer Manag Res. 2018;10:4517–22.
Henricks LM, Siemerink EJM, Rosing H, Meijer J, Goorden SMI, Polstra AM, et al. Capecitabine-based treatment of a patient with a novel DPYD genotype and complete dihydropyrimidine dehydrogenase deficiency. Int J Cancer. 2018;142:424–30.
Lazar A, Mau-Holzmann UA, Kolb H, Reichenmiller HE, Riess O, Schomig E. Multiple organ failure due to 5-fluorouracil chemotherapy in a patient with a rare dihydropyrimidine dehydrogenase gene variant. Onkologie. 2004;27:559–62.
Ly RC, Schmidt RE, Kiel PJ, Pratt VM, Schneider BP, Radovich M. et al. Severe Capecitabine Toxicity Associated With a Rare DPYD Variant Identified Through Whole-Genome Sequencing. JCO Precis Oncol. 2020;4:632–8.
Vilquin P, Medard Y, Thomas F, Goldwirt L, Teixeira L, Mourah S, et al. DPYD genotype should be extended to rare variants: report on two cases of phenotype / genotype discrepancy. Cancer Chemother Pharm. 2025;95:16.
Knikman JE, Lopez-Yurda M, Meulendijks D, Deenen MJ, Schellens JHM, Beijnen J, et al. A Nomogram to Predict Severe Toxicity in DPYD Wild-Type Patients Treated With Capecitabine-Based Anticancer Regimens. Clin Pharm Ther. 2024;115:269–77.
Le Teuff G, Cozic N, Boyer JC, Boige V, Diasio RB, Taieb J, et al. Dihydropyrimidine dehydrogenase gene variants for predicting grade 4-5 fluoropyrimidine-induced toxicity: FUSAFE individual patient data meta-analysis. Br J Cancer. 2024;130:808–18.
De Mattia E, Polesel J, Scarabel L, Cecchin E. Clinical implications of a gain-of-function genetic polymorphism in DPYD (rs4294451) in colorectal cancer patients treated with fluoropyrimidines. Front Pharm. 2024;15:1516375.
Xiao Q, Zhou Y, Lauschke VM. Ethnogeographic and inter-individual variability of human ABC transporters. Hum Genet. 2020;139:623–46.
Kugimiya N, Nishimoto A, Hosoyama T, Ueno K, Enoki T, Li TS, et al. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J Cell Mol Med. 2015;19:1569–81.
Wilson BJ, Schatton T, Zhan Q, Gasser M, Ma J, Saab KR, et al. ABCB5 identifies a therapy-refractory tumor cell population in colorectal cancer patients. Cancer Res. 2011;71:5307–16.
Moitra K, Scally M, McGee K, Lancaster G, Gold B, Dean M. Molecular evolutionary analysis of ABCB5: the ancestral gene is a full transporter with potentially deleterious single nucleotide polymorphisms. PLoS ONE. 2011;6:e16318.
Chen Q, Meng F, Wang L, Mao Y, Zhou H, Hua D, et al. A polymorphism in ABCC4 is related to efficacy of 5-FU/capecitabine-based chemotherapy in colorectal cancer patients. Sci Rep. 2017;7:7059.
Hagmann W, Jesnowski R, Faissner R, Guo C, Lohr JM. ATP-binding cassette C transporters in human pancreatic carcinoma cell lines. Upregulation 5-fluorouracil-resistant Cells Pancreatol. 2009;9:136–44.
Zhang G, Wang Z, Qian F, Zhao C, Sun C. Silencing of the ABCC4 gene by RNA interference reverses multidrug resistance in human gastric cancer. Oncol Rep. 2015;33:1147–54.
Cecchin E, De Mattia E, Ecca F, Toffoli G. Host genetic profiling to increase drug safety in colorectal cancer from discovery to implementation. Drug Resist Updat. 2018;39:18–40.
De Mattia E, Cecchin E, Toffoli G. Pharmacogenomics of intrinsic and acquired pharmacoresistance in colorectal cancer: Toward targeted personalized therapy. Drug Resist Updat. 2015;20:39–70.
Sethy C, Kundu CN. 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed Pharmacother. 2021;137:111285.
Wyatt MD, Wilson DM. 3rd. Participation of DNA repair in the response to 5-fluorouracil. Cell Mol Life Sci. 2009;66:788–99.
Mini E, Landini I, Lucarini L, Lapucci A, Napoli C, Perrone G, et al. The Inhibitory Effects of HYDAMTIQ, a Novel PARP Inhibitor, on Growth in Human Tumor Cell Lines With Defective DNA Damage Response Pathways. Oncol Res. 2017;25:1441–51.
Paul S, Chatterjee S, Sinha S, Dash SR, Pradhan R, Das B, et al. Veliparib (ABT-888), a PARP inhibitor potentiates the cytotoxic activity of 5-fluorouracil by inhibiting MMR pathway through deregulation of MSH6 in colorectal cancer stem cells. Expert Opin Ther Targets. 2023;27:999–1015.
Wang F, Gouttia OG, Wang L, Peng A. PARP1 Upregulation in Recurrent Oral Cancer and Treatment Resistance. Front Cell Dev Biol. 2021;9:804962.
Du L, Xiong T, He Q, Wang Y, Shen J, Peng Y, et al. The Thr241Met polymorphism in the XRCC3 gene is associated with increased risk of cancer in Chinese mainland populations. Tumour Biol. 2014;35:1371–6.
Qin LY, Chen X, Li P, Yang Z, Mo WN. Association between the XRCC3 Thr241Met polymorphism and cervical cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2014;14:6703–7.
Liu Y, Chen H, Chen L, Hu C. Prediction of genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 in the survival of colorectal cancer receiving chemotherapy in the Chinese population. Hepatogastroenterology. 2012;59:977–80.
Ott K, Rachakonda PS, Panzram B, Keller G, Lordick F, Becker K, et al. DNA repair gene and MTHFR gene polymorphisms as prognostic markers in locally advanced adenocarcinoma of the esophagus or stomach treated with cisplatin and 5-fluorouracil-based neoadjuvant chemotherapy. Ann Surg Oncol. 2011;18:2688–98.
Cecchin E, De Mattia E, Toffoli G. Nuclear receptors and drug metabolism for the personalization of cancer therapy. Expert Opin Drug Metab Toxicol. 2016;12:291–306.
De Mattia E, Cecchin E, Roncato R, Toffoli G. Pregnane X receptor, constitutive androstane receptor and hepatocyte nuclear factors as emerging players in cancer precision medicine. Pharmacogenomics. 2016;17:1547–71.
De Mattia E, Dreussi E, Cecchin E, Toffoli G. Pharmacogenetics of the nuclear hormone receptors: the missing link between environment and drug effects?. Pharmacogenomics. 2013;14:2035–54.
Weikum ER, Liu X, Ortlund EA. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018;27:1876–92.
Abu N, Othman N, Nazarie KWH, Jamal WF. R. Integrative meta-analysis for the identification of hub genes in chemoresistant colorectal cancer. Biomark Med. 2020;14:525–37.
Hagos Y, Wegner W, Kuehne A, Floerl S, Marada VV, Burckhardt G, et al. HNF4alpha induced chemosensitivity to oxaliplatin and 5-FU mediated by OCT1 and CNT3 in renal cell carcinoma. J Pharm Sci. 2014;103:3326–34.
Jiang H, Chen K, He J, Pan F, Li J, Chen J, et al. Association of pregnane X receptor with multidrug resistance-related protein 3 and its role in human colon cancer chemoresistance. J Gastrointest Surg. 2009;13:1831–8.
Schockel L, Woischke C, Surendran SA, Michl M, Schiergens T, Holscher A, et al. PPARG activation promotes the proliferation of colorectal cancer cell lines and enhances the antiproliferative effect of 5-fluorouracil. BMC Cancer. 2024;24:234.
Yu J, Chen M, Sang Q, Li F, Xu Z, Yu B, et al. Super-enhancer Activates Master Transcription Factor NR3C1 Expression and Promotes 5-FU Resistance in Gastric Cancer. Adv Sci. 2025;12:e2409050.
Chen L, Wen A. Unveiling the role of O(6)-methylguanine-DNA methyltransferase in cancer therapy: insights into alkylators, pharmacogenomics, and others. Front Oncol. 2024;14:1424797.
Bignucolo A, Scarabel L, Toffoli G, Cecchin E, De Mattia E. Predicting drug response and toxicity in metastatic colorectal cancer: the role of germline markers. Expert Rev Clin Pharm. 2022;15:689–713.
Park JH, Kim NS, Park JY, Chae YS, Kim JG, Sohn SK, et al. MGMT -535G>T polymorphism is associated with prognosis for patients with metastatic colorectal cancer treated with oxaliplatin-based chemotherapy. J Cancer Res Clin Oncol. 2010;136:1135–42.
Dong Y, Wang Z, Xie GF, Li C, Zuo WW, Meng G, et al. Pregnane X receptor is associated with unfavorable survival and induces chemotherapeutic resistance by transcriptional activating multidrug resistance-related protein 3 in colorectal cancer. Mol Cancer. 2017;16:71.
Lancaster CS, Sprowl JA, Walker AL, Hu S, Gibson AA, Sparreboom A. Modulation of OATP1B-type transporter function alters cellular uptake and disposition of platinum chemotherapeutics. Mol Cancer Ther. 2013;12:1537–44.
Theile D, Grebhardt S, Haefeli WE, Weiss J. Involvement of drug transporters in the synergistic action of FOLFOX combination chemotherapy. Biochem Pharm. 2009;78:1366–73.
De Mattia E, Roncato R, Dalle Fratte C, Ecca F, Toffoli G, Cecchin E. The use of pharmacogenetics to increase the safety of colorectal cancer patients treated with fluoropyrimidines. Cancer Drug Resist. 2019;2:116–30.
Maslarinou A, Manolopoulos VG, Ragia G. Pharmacogenomic-guided dosing of fluoropyrimidines beyond DPYD: time for a polygenic algorithm?. Front Pharm. 2023;14:1184523.
De Mattia E, Cecchin E, Montico M, Labriet A, Guillemette C, Dreussi E, et al. Association of STAT-3 rs1053004 and VDR rs11574077 With FOLFIRI-Related Gastrointestinal Toxicity in Metastatic Colorectal Cancer Patients. Front Pharm. 2018;9:367.
De Mattia E, Polesel J, Roncato R, Labriet A, Bignucolo A, Dreussi E, et al. Germline Polymorphisms in the Nuclear Receptors PXR and VDR as Novel Prognostic Markers in Metastatic Colorectal Cancer Patients Treated With FOLFIRI. Front Oncol. 2019;9:1312.
De Mattia E, Toffoli G, Polesel J, D’Andrea M, Corona G, Zagonel V, et al. Pharmacogenetics of ABC and SLC transporters in metastatic colorectal cancer patients receiving first-line FOLFIRI treatment. Pharmacogenet Genomics. 2013;23:549–57.
Labriet A, De Mattia E, Cecchin E, Levesque E, Jonker D, Couture F, et al. Improved Progression-Free Survival in Irinotecan-Treated Metastatic Colorectal Cancer Patients Carrying the HNF1A Coding Variant p.I27L. Front Pharm. 2017;8:712.
Russell LE, Zhou Y, Almousa AA, Sodhi JK, Nwabufo CK, Lauschke VM. Pharmacogenomics in the era of next generation sequencing - from byte to bedside. Drug Metab Rev. 2021;53:253–78.
Gong L, Klein CJ, Caudle KE, Moyer AM, Scott SA, Whirl-Carrillo M, et al. Integrating Pharmacogenomics into the Broader Construct of Genomic Medicine: Efforts by the ClinGen Pharmacogenomics Working Group (PGxWG). Clin Chem. 2025;71:36–44.
Zhou Y, Lauschke VM. Population pharmacogenomics: an update on ethnogeographic differences and opportunities for precision public health. Hum Genet. 2022;141:1113–36.
Zhou Y, Park Y, Camara MD, Lauschke VM. Opportunities and Challenges of Population Pharmacogenomics. Ann Hum Genet. 2025;89:384–97.
Funding
This work was supported by the Italian Ministry of Health (Ricerca Corrente) and European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 668353. We also acknowledge funding from the Swedish Research Council [grant numbers: 2021-02801 and 2023-03015], by the SciLifeLab and Wallenberg National Program for Data-Driven Life Science [WASPDDLS22:006], Cancerfonden [23-0763PT and 24-3735Pj], the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2180—390900677, the Robert Bosch Foundation, Stuttgart, Germany, and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2023-00209528).
Author information
Authors and Affiliations
Contributions
EDM, EC and VML conceptualized and designed the study. MS and FP provided and collected clinical samples and recorded clinical data. EDM directed and supervised the execution of sequencing experiments. EP, RR, and LS carried out sample processing and sequencing experiments. JP created the dataset by selecting matched cases and controls. YP and YZ carried out bio-informatic data processing and computational analysis. EDM, YP, VML and EC carried out interpretation of data. EDM and YP wrote the manuscript. EC, VML, GT, MS, HJG and JJS edited and revised the manuscript. EC and VML supervised the overall study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
VML is co-founder, CEO and shareholder of HepaPredict AB, as well as co-founder and shareholder of Shanghai Hepo Biotechnology Ltd. All other authors declare that they have no competing interests.
Ethics approval and consent to participate
The study population was selected from the prospective PREPARE trial (NCT03093818, ClinicalTrials.gov). The PREPARE study adhered to the principles outlined in the 1975 Declaration of Helsinki (with the 1983 revision) and was granted ethical approval by the local ethic committee. All patients provided written informed consent before entering the study to donate their anonymized blood samples for further studies including NGS analysis. All experiments were carried out in accordance with the relevant guidelines and regulations of Centro di Riferimento Oncologico di Aviano.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
De Mattia, E., Park, Y., Peruzzi, E. et al. Integration of germline pharmacogenomic burden to predict fluoropyrimidine-related toxicity – A secondary analysis of the PREPARE trial. Oncogene 44, 4352–4362 (2025). https://doi.org/10.1038/s41388-025-03587-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03587-7