Abstract
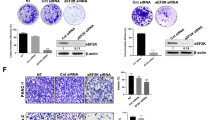
Pancreatic ductal adenocarcinoma (PDAC) has two subtypes: the “classical/progenitor” type and “basal-like/squamous” type, the latter of which has poor clinical outcomes with no effective treatment strategies. We aimed to elucidate the role of epithelial membrane protein 1 (EMP1) in PDAC and its potential as a therapeutic target, particularly in aggressive disease such as “basal-like/squamous” type of PDAC. We examined the association of EMP1 expression using patient-derived organoids (PDOs) of human PDAC, K-RASLSL-G12D, Trp 53LSL-R172H, and Pdx1-Cre recombinase mice, human PDAC cell lines, and publicly available clinical datasets. The functional roles of EMP1 were evaluated in vitro and in vivo through its knockout and stable overexpression. EMP1 knockout reduced proliferation, metastasis, and drug resistance, whereas overexpression enhanced malignant features. Transcriptomic analysis revealed that EMP1 promotes epithelial-mesenchymal transition (EMT), extracellular matrix remodeling, and the K-RAS signaling pathway. EMP1 expression is inversely implicated in the oxidative phosphorylation pathway, which is characteristic of the “classical/progenitor” type. Furthermore, integrated analysis revealed an association between EMP1 expression and ERK phosphorylation. EMP1 plays a crucial role in the pathogenesis of PDAC, as it contributes to the proliferative and metastatic characteristics of PDAC. This study suggests that EMP1 may be a potential therapeutic target gene for aggressive disease.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data pertaining to this study are accessible from the corresponding authors upon reasonable request.
References
Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics 2025. CA Cancer J Clin. 2025;75:10–45.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78.
Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3.
Chan-Seng-Yue M, Kim JC, Wilson GW, Wilson GW, Ng K, Figueroa EF, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40.
Miyabayashi K, Baker LA, Deschênes A, Raub B, Caligiuri G, et al. Intraductal transplantation models of human pancreatic ductal adenocarcinoma reveal progressive transition of molecular subtypes. Cancer Discov. 2020;10:1566–89.
Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184:6119–37.e26.
Matsumoto K, Fujimori N, Ichihara K, Takeno A, Murakami M, Ohno A, et al. Patient-derived organoids of pancreatic ductal adenocarcinoma for subtype determination and clinical outcome prediction. J Gastroenterol. 2024;59:629–40.
Romero-Calvo I, Weber CR, Ray M, Brown M, Kirby K, Nandi RK, et al. Human organoids share structural and genetic features with primary pancreatic adenocarcinoma tumors. Mol Cancer Res. 2019;17:70–83.
Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–67.e6.
Cañellas-Socias A, Cortina C, Hernando-Momblona X, Palomo-Ponce S, Mulholland EJ, Turon G, et al. Metastatic recurrence in colorectal cancer arises from residual EMP1+ cells. Nature. 2022;611:603–13.
Wang Y, Zhang L, Yao C, Ma Y, Liu Y. Epithelial membrane protein 1 promotes sensitivity to RSL3-induced ferroptosis and intensifies gefitinib resistance in head and neck cancer. Oxidative Med Cell Longev. 2022;2022:4750671.
Ahmat Amin MKB, Shimizu A, Zankov DP, Sato A, Kurita S, Ito M, et al. Epithelial membrane protein 1 promotes tumor metastasis by enhancing cell migration via copine-III and Rac1. Oncogene. 2018;37:5416–34.
Miao L, Jiang Z, Wang J, Yang N, Qi Q, Zhou W, et al. Epithelial membrane protein 1 promotes glioblastoma progression through the PI3K/AKT/mTOR signaling pathway. Oncol Rep. 2019;42:605–14.
Liu Y, Ding Y, Nie Y, Yang M. EMP1 promotes the proliferation and invasion of ovarian cancer cells through activating the MAPK pathway. Onco Targets Ther. 2020;13:2047–55.
Sato A, Rahman NIA, Shimizu A, Ogita H. Cell-to-cell contact-mediated regulation of tumor behavior in the tumor microenvironment. Cancer Sci. 2021;112:4005–12.
Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83.
Kawano T, Murata M, Kang JH, Piao JS, Narahara S, Hyodo F, et al. Ultrasensitive MRI detection of spontaneous pancreatic tumors with nanocage-based targeted contrast agent. Biomaterials. 2018;152:37–46.
Duan X, Zhang T, Feng L, de Silva N, Greenspun B, Wang X, et al. A pancreatic cancer organoid platform identifies an inhibitor specific to mutant KRAS. Cell Stem Cell. 2024;31:71–88.e8.
Hu WM, Liu SQ, Zhu KF, Li W, Yang ZJ, Yang Q, et al. The ALOX5 inhibitor Zileuton regulates tumor-associated macrophage M2 polarization by JAK/STAT and inhibits pancreatic cancer invasion and metastasis. Int Immunopharmacol. 2023;121:110505.
Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. 2018;45:681–695.e4.
Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25:628–40.
Wang YW, Cheng HL, Ding YR, Chou LH, Chow NH. EMP1, EMP2, and EMP3 as novel therapeutic targets in human cancer. Biochim Biophys Acta Rev Cancer. 2017;1868:199–211.
Ahmat Amin MKB, Shimizu A, Ogita H. The pivotal roles of the epithelial membrane protein family in cancer invasiveness and metastasis. Cancers. 2019;11:1–21.
Liu W, Li J, Zhao R, Lu Y, Huang P. The uridine diphosphate (UDP)-glycosyltransferases (UGTs) superfamily: the role in tumor cell metabolism. Front Oncol. 2023;12:1088458.
Hu DG, Meech R, McKinnon RA, Mackenzie PI. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug Metab Rev. 2014;46:421–58.
Sheng B, Jiang Y, Wu D, Lai N, Ye Z, Zhang B, et al. RNAi-mediated SYT14 knockdown inhibits the growth of human glioma cell line U87MG. Brain Res Bull. 2018;140:60–4.
Yoo HB, Moon JW, Kim HR, Lee HS, Miyabayashi K, Park CH, et al. A TEAD2-driven endothelial-like program shapes basal-like differentiation and metastasis of pancreatic cancer. Gastroenterology. 2023;165:133–148.e17.
Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 2016;14:2281–8.
Low RRJ, Fung KY, Gao H, Preaudet A, Dagley LF, Yousef J, et al. S100 family proteins are linked to organoid morphology and EMT in pancreatic cancer. Cell Death Differ. 2023;30:1155–65.
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
Bouzid H, Soualmia F, Oikonomopoulou K, Soosaipillai A, Walker F, Louati K, et al. Kallikrein-related peptidase 6 (KLK6) as a contributor toward an aggressive cancer cell phenotype: a potential role in colon cancer peritoneal metastasis. Biomolecules. 2022;12:1003.
Simard M, Grenier A, Rioux G, Tremblay A, Blais I, Flamand N, et al. Remodeling of the dermal extracellular matrix in a tissue-engineered psoriatic skin model by n-3 polyunsaturated fatty acids. Biomedicines. 2022;10:1078.
Spada S, Tocci A, Di Modugno F, Nisticò P. Fibronectin as a multiregulatory molecule crucial in tumor matrisome: from structural and functional features to clinical practice in oncology. J Exp Clin Cancer Res. 2021;40:102.
Maurer C, Holmstrom SR, He J, Laise P, Su T, Ahmed A, et al. Experimental microdissection enables functional 21armonization of pancreatic cancer subtypes. Gut. 2019;68:1034–43.
Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25:1741–55.e7.
Araki O, Tsuda M, Omatsu M, Namikawa M, Sono M, Fukunaga Y, et al. Brg1 controls stemness and metastasis of pancreatic cancer through regulating hypoxia pathway. Oncogene. 2023;42:2139–52.
Liu S, Shi J, Wang L, Huang Y, Zhao B, Ding H, et al. Loss of EMP1 promotes the metastasis of human bladder cancer cells by promoting migration and conferring resistance to ferroptosis through activation of PPAR gamma signaling. Free Radic Biol Med. 2022;189:42–57.
Wang HT, Kong JP, Ding F, Wang XQ, Wang MR, Liu LX, et al. Analysis of gene expression profile induced by EMP-1 in esophageal cancer cells using cDNA microarray. World J Gastroenterol. 2003;9:392–8.
Sun G, Zhao G, Lu Y, Wang Y, Yang C. Association of EMP1 with gastric carcinoma invasion, survival and prognosis. Int J Oncol. 2014;45:1091–8.
Kordshouli SO, Tahmasebi A, Moghadam A, Ramezani A, Niazi A. A comprehensive meta-analysis of transcriptome data to identify signature genes associated with pancreatic ductal adenocarcinoma. PLoS ONE. 2024;19:e0289561.
Wang X, Jiang S, Zhou X, Wang X, Li L, Tang J. Prognostic-related genes for pancreatic cancer typing and immunotherapy response prediction based on single-cell sequencing data and bulk sequencing data. Oncol Res. 2023;31:697–714.
Yang J, Liu Y, Liu S. The role of epithelial-mesenchymal transition and autophagy in pancreatic ductal adenocarcinoma invasion. Cell Death Dis. 2023;14:506.
Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84.
Ni Q, Zhang Y, Tao R, Li X, Zhu J. MicroRNA-95-3p serves as a contributor to cisplatin resistance in human gastric cancer cells by targeting EMP1/PI3K/AKT signaling. Aging. 2021;13:8665–87.
Li J, Liu J, Wang H, Ma J, Wang Y, Xu W. Single-cell analyses EMP1 as a marker of the ratio of M1/M2 macrophages is associated with EMT, immune infiltration, and prognosis in bladder cancer. Bladder. 2023;10:e21200011.
Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178:160–175.e27.
Gabitova-Cornell L, Surumbayeva A, Peri S, Franco-Barraza J, Restifo D, Weitz N, et al. Cholesterol pathway inhibition induces TGF-beta signaling to promote basal differentiation in pancreatic cancer. Cancer Cell. 2020;38:567–83.e11.
Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–14.
Guillemette C, Lévesque É, Rouleau M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin Pharmacol Ther. 2014;96:324–39.
Van Sciver RE, Lee MP, Lee CD, Lafever AC, Svyatova E, Kanda K, et al. A new strategy to control and eradicate “Undruggable” oncogenic K-RAS-driven pancreatic cancer: molecular insights and core principles learned from developmental and evolutionary biology. Cancers. 2018;10:142.
Funding
This study received partial funding from the Mochida Memorial Foundation for Medical and Pharmaceutical Research (GAKF800717), which awarded a grant to NF. This study was also supported by Nanken-Kyoten (Grant No. 2025-kokunai 37), Science Tokyo (awarded to YO). Additional support was received from the Daiwa Securities Foundation (GAKF800710), (awarded to NF), as well as KAKENHI grants from the Japan Society for the Promotion of Science (JSPS) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan, granted to AT (JP20K17023), YO (JP22H04993), KU (JP23K15013), and K.T. (JP23K19514), and NF (JP25K11254).
Author information
Authors and Affiliations
Contributions
AO collected and curated the data, designed the study methodology, was involved with the provision of study materials and the preparation of the data presentation, conceptualized the study, analyzed and validated the data, wrote the original draft of the manuscript, and contributed to reviewing and editing the manuscript. NF supervised the experiment, acquired funding, and reviewed and edited the manuscript. KM developed the study methodology, curated the data, and was involved with the provision of study materials. OS developed the study methodology, curated the data, and was involved with the provision of study materials. AT developed the study methodology, acquired funding, and was involved with the provision of study materials. TY was involved in the provision of study materials. AS was also involved in the provision of study materials. SK developed the study methodology, curated the data, and was involved with the provision of study materials. TU was involved with the provision of study materials. MM was involved with the provision of study materials. KT acquired funding and was involved with the provision of study materials. KU acquired funding and was involved with the provision of study materials. TO developed the study methodology and was involved with the provision of study materials. SH developed the study methodology and contributed to the review and editing of the manuscript. Yoshinao Oda was instrumental in developing the study methodology and providing study materials. KIN developed the study methodology and was involved with the provision of study materials. Yoshihiro Ogawa supervised the experiment, acquired funding, and reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The clinical data, tumor samples, and PDOs from consecutive cases of PDAC were prospectively obtained at Kyushu University Hospital between April 2023 and March 2024. This study followed the ethical standards of the 1964 Declaration of Helsinki and received approval from the Ethics Committee of Kyushu University Hospital (Approval no. 22121-02 and 22161-01). All participants provided written informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ohno, A., Fujimori, N., Matsumoto, K. et al. Role of a transmembrane protein, epithelial membrane protein 1, in the pathogenesis of pancreatic ductal adenocarcinoma. Oncogene 45, 307–321 (2026). https://doi.org/10.1038/s41388-025-03633-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03633-4