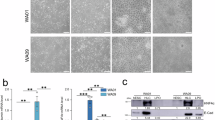
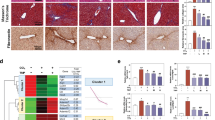
Abstract
Activating mutations in the ligand-binding domain of the estrogen receptor (ER)-encoding (ESR1) gene are present in up to 40% of metastatic breast cancer (BC) patients and are strongly associated with a high risk of liver metastasis (LM) formation. Using the MCF-7 BC model, we investigated whether the increased hepatic tropism of ESR1-mutated BC cells is driven by their metabolic adaptation to the liver microenvironment. Indeed, metabolomic analysis revealed elevated metabolites related to the urea cycle (UC) in LM-forming ESR1-mutated cells compared to wild-type (WT) ER-expressing cells, which failed to generate LM. The subsequent proteomic, western blotting, and qPCR analyses demonstrated a dramatic upregulation of the UC constituent, the mitochondrial ornithine/citrulline transporter SLC25A15, in liver-predilected ESR1-mutated cells relative to their WT counterpart cells. Unlike WT cells, ESR1-mutated cells readily formed spheroids and exhibited enhanced migration in liver mimicking hepatocyte-conditioned media. In addition, we employed a novel ex vivo approach where ESR1 mutated cells were seeded onto colonized fresh liver tissue—which was abolished by SLC25A15 knockout. Moreover, SLC25A15 knockout robustly reduced the ability of ESR1-mutated cells to establish LM in vivo. These findings highlight SLC25A15-mediated dysregulation of the UC as a critical driver of BC hepatic metastasis and identify SLC25A15 as a potential therapeutic target for disrupting metastatic spread of BC to the liver.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
References
Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol 2022;95. https://doi.org/10.1259/BJR.20211033.
Pu CC, Yin L, Yan JM. Risk factors and survival prediction of young breast cancer patients with liver metastases: a population-based study. Front Endocrinol 2023; 14. https://doi.org/10.3389/fendo.2023.1158759.
Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73:6856–64.
Zinger L, Merenbakh-Lamin K, Klein A, Elazar A, Journo S, Boldes T, et al. Ligand-binding domain–activating mutations of ESR1 rewire cellular metabolism of breast cancer cells. Clin Cancer Res. 2019;25:2900–14. https://doi.org/10.1158/1078-0432.ccr-18-1505.
Pegram M, Jackisch C, Johnston SRD. Estrogen/HER2 receptor crosstalk in breast cancer: combination therapies to improve outcomes for patients with hormone receptor-positive/HER2-positive breast cancer. npj Breast Cancer. 2023;9:45. https://doi.org/10.1038/s41523-023-00533-2.
Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67.
Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–83. https://doi.org/10.1038/nrclinonc.2015.117.
Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 mutations in cell-free DNA and Outcomes in Metastatic Breast Cancer. JAMA Oncol. 2016;2:1310. https://doi.org/10.1001/jamaoncol.2016.1279.
Reinert T, Coelho GP, Mandelli J, Zimermann E, Zaffaroni F, Bines J, et al. Association of ESR1 mutations and visceral metastasis in patients with estrogen receptor-positive advanced breast cancer from Brazil. J Oncol. 2019;2019:1–5. https://doi.org/10.1155/2019/1947215.
Wu Y, Li Z, Lee AV, Oesterreich S, Luo B. Liver tropism of ER mutant breast cancer is characterized by unique molecular changes and immune infiltration. Breast Cancer Res Treat. 2024;205:371–86. https://doi.org/10.1007/s10549-024-07255-4.
Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XHF. Metastasis Organotropism: redefining the congenial soil. Dev Cell. 2019;49:375–91. https://doi.org/10.1016/j.devcel.2019.04.012.
Hajaj E, Sciacovelli M, Frezza C, Erez A. The context-specific roles of urea cycle enzymes in tumorigenesis. Mol Cell. 2021;81:3749–59.
Keshet R, Szlosarek P, Carracedo A, Erez A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat Rev Cancer. 2018;18:634–45.
Qiu F, Chen YR, Liu X, Chu CY, Shen LJ, Xu J, et al. Cancer: Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci Signal 2014, 7. https://doi.org/10.1126/scisignal.2004761.
Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015;527:379–83. https://doi.org/10.1038/nature15529.
Ji S. Overexpression of SLC25A15 is involved in the proliferation of cutaneous melanoma and leads to poor prognosis. Medecine/Sci. 2018;34:74–80. https://doi.org/10.1051/medsci/201834f113.
Zou Z, Hu X, Luo T, Ming Z, Chen X, Xia L, et al. Naturally-occurring spinosyn A and its derivatives function as argininosuccinate synthase activator and tumor inhibitor. Nat Commun. 2021;12:2263. https://doi.org/10.1038/s41467-021-22235-8.
Zhang T, Hu L, Tang JF, Xu H, Tian K, Wu MN, et al. Metformin inhibits the urea cycle and reduces putrescine generation in colorectal cancer cell lines. Molecules. 2021;26:1990. https://doi.org/10.3390/molecules26071990.
Taya M, Merenbakh-Lamin K, Zubkov A, Honig Z, Kurolap A, Mayer O, et al. Beyond endocrine resistance: estrogen receptor (ESR1) activating mutations mediate chemotherapy resistance through the JNK/c-Jun MDR1 pathway in breast cancer. Breast Cancer Res Treat. 2024;209:431–49.
Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–7. https://doi.org/10.1038/nmeth.1314.
Feldman B, Fedida-Metula S, Nita J, Sekler I, Fishman D. Coupling of mitochondria to store-operated Ca(2+)-signaling sustains constitutive activation of protein kinase B/Akt and augments survival of malignant melanoma cells. Cell Calcium. 2010;47:525–37. https://doi.org/10.1016/j.ceca.2010.05.002.
Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67. https://doi.org/10.1158/1078-0432.CCR-13-2332.
Wei Q, Qian Y, Yu J, Wong CC. Metabolic rewiring in the promotion of cancer metastasis: mechanisms and therapeutic implications. Oncogene. 2020;39:6139–56. https://doi.org/10.1038/s41388-020-01432-7.
Häussinger D, Sies H, Gerok W. Functional hepatocyte heterogeneity in ammonia metabolism. The intercellular glutamine cycle. J Hepatol. 1985;1:3–14. https://doi.org/10.1016/S0168-8278(85)80063-5.
Martí i Líndez AA, Reith W. Arginine-dependent immune responses. Cell Mol Life Sci. 2021;78:5303–24. https://doi.org/10.1007/s00018-021-03828-4.
Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7:277–87. https://doi.org/10.1158/2159-8290.CD-15-1523.
Corné J, Quillien V, Callens C, Portois P, Bidard FC, Jeannot E, et al. Development of sensitive and robust multiplex digital PCR assays for the detection of ESR1 mutations in the plasma of metastatic breast cancer patients. Clin Chim Acta. 2023;545:117366. https://doi.org/10.1016/j.cca.2023.117366.
Decotret LR, Shi R, Thomas KN, Hsu M, Pallen CJ, Bennewith KL. Development and validation of an advanced ex vivo brain slice invasion assay to model glioblastoma cell invasion into the complex brain microenvironment. Front Oncol 2023;13. https://doi.org/10.3389/fonc.2023.976945.
Neve A, Santhana Kumar K, Tripolitsioti D, Grotzer MA, Baumgartner M. Investigation of brain tissue infiltration by medulloblastoma cells in an ex vivo model. Sci Rep. 2017;7. 5297. https://doi.org/10.1038/s41598-017-05573-w.
Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P, Peterziel H. An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer. 2018;18:103. https://doi.org/10.1186/s12885-018-4007-4.
Grotzer MA, Neve A, Baumgartner M. Dissecting brain tumor growth and metastasis in vitro and ex vivo. J Cancer Metastasis Treat. 2016;2:149. https://doi.org/10.20517/2394-4722.2016.02.
Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016;4. https://doi.org/10.3389/fbioe.2016.00012.
Dupuy F, Tabariès S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–89. https://doi.org/10.1016/j.cmet.2015.08.007.
Ghosh N, Mahalanobish S. PCS. Reprogramming of urea cycle in cancer: Mechanism, regulation and prospective therapeutic scopes. Biochem Pharm. 2024;228:116326 https://doi.org/10.1016/j.bcp.2024.116326.
Dermit M, Dodel M, Mardakheh FK. Methods for monitoring and measurement of protein translation in time and space. Mol Biosyst. 2017;13:2477–88. https://doi.org/10.1039/c7mb00476a.
Zhang Q, Wei T, Jin W, Yan L, Shi L, Zhu S, et al. Deficiency in SLC25A15, a hypoxia-responsive gene, promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J Hepatol. 2024;80:293–308. https://doi.org/10.1016/j.jhep.2023.10.024.
Goddard ET, Fischer J, Schedin P. A portal vein injection model to study liver metastasis of breast cancer. J Vis Exp 2016;2016. https://doi.org/10.3791/54903.
Khanna C. Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2004;26:513–23. https://doi.org/10.1093/carcin/bgh261.
Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, et al. Mitochondria ‘fuel’ breast cancer metabolism: Fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle. 2012;11:4390–401. https://doi.org/10.4161/cc.22777.
Lytovchenko O, Kunji ERS. Expression and putative role of mitochondrial transport proteins in cancer. Biochim Biophys Acta - Bioenerg. 2017;1858:641–54. https://doi.org/10.1016/j.bbabio.2017.03.006.
Zuo Q, Mogol AN, Liu YJ, Santaliz Casiano A, Chien C, Drnevich J, et al. Targeting metabolic adaptations in the breast cancer–liver metastatic niche using dietary approaches to improve endocrine therapy efficacy. Mol Cancer Res. 2022;20:923–37. https://doi.org/10.1158/1541-7786.MCR-21-0781.
Funding
This study was supported by the Israel Science Foundation to D.F, I.W and T.R (grant no 2861/24), and by The Dubrovsky Family Research Fund for Solid Cancers, the Cancer Biology Research Center, Tel-Aviv University to I.W and T.R.
Author information
Authors and Affiliations
Contributions
MT- Collected the data, performed the analysis of in vitro and in vivo assays, and wrote the manuscript. DF- Conceived the research, collected the data, and performed the analysis of in vitro assays. FK- performed in vivo experiment. IS- Conceived and contributed to the analysis. IW- Conceived and designed the analysis. TR- Conceived and designed the analysis, performed the analysis, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing of interest.
Ethical approval
The experiments on animals were conducted in accordance with the institutional guidelines of the Sourasky Medical Center, in accordance with current regulations and standards of the institution's Animal Care and Use Committee.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Taya, M., Fishman, D., Kanani, F. et al. The role of the SLC25A15 transporter in the formation of liver metastasis in ESR1-mutated breast cancer. Oncogene 45, 603–619 (2026). https://doi.org/10.1038/s41388-025-03640-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03640-5