Abstract
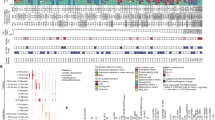
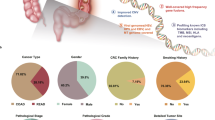
Colorectal cancer (CRC) cell lines represent the main molecular subtypes of tumors and are valuable models for preclinical investigations. However, cell lines can diverge over time and careful selection of models based on their molecular features is key. We have authenticated 103 commonly used CRC cell lines and present the mutation profiles of 20 CRC-relevant genes sequenced to an average depth of 575 times coverage. The cell lines reflected the distinct mutation patterns of hypermutation phenotypes associated with microsatellite instability and pathogenic POLE mutations. Hypermutated cell lines appeared to have a stronger mutational divergence and more frequent subclonal mutations, while mutations not associated with hypermutation were more frequently homozygous or hemizygous, classified as pathogenic, and subject to stronger selection pressure. Loss of heterozygosity at mutated loci was primarily observed in tumor suppressor genes. Genetic interactions based on co-occurring mutations identified cell lines representative of particularly aggressive subtypes of CRC, including concurrent BRAF p.V600 and truncating APC mutations, as well as APC/TP53/RAS triple mutations with double hits of APC. This study provides a resource to guide the selection of cell lines for functional studies of CRC, and detailed mutation data including classifications of pathogenicity, variant allele frequencies and illustrations of the mutation distribution along the length of encoded proteins are included.
Similar content being viewed by others
Data availability
All mutations (processed data) and STR profiles are available in the supplementary material.
Code availability
The MiniCN R package, including source code, example data, and documentation, is freely available at https://github.com/SveenLab/miniCN. Additional computer code and supporting information used for data processing and plotting are accessible on Zenodo: https://doi.org/10.5281/zenodo.17357561.
References
Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–6.
Geraghty RJ, Capes-Davis A, Davis JM, Downward J, Freshney RI, Knezevic I, et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 2014;111:1021–46.
Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumagi A, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24:794–806.
Buikhuisen JY, Gomez Barila PM, Cameron K, Suijkerbuijk SJE, Lieftink C, di Franco S, et al. Subtype-specific kinase dependency regulates growth and metastasis of poor-prognosis mesenchymal colorectal cancer. J Exp Clin Cancer Res. 2023;42:56.
McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007;104:19936–41.
Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8.
Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med. 2019;381:1632–43.
Tan L, Tran B, Tie J, Markman B, Ananda S, Tebbutt NC, et al. A Phase Ib/II Trial of Combined BRAF and EGFR Inhibition in BRAF V600E Positive Metastatic Colorectal Cancer and Other Cancers: The EVICT (Erlotinib and Vemurafenib In Combination Trial) Study. Clin Cancer Res. 2023;29:1017–30.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Jin H, Zhang C, Zwahlen M, von Feilitzen K, Karlsson M, Shi M, et al. Systematic transcriptional analysis of human cell lines for gene expression landscape and tumor representation. Nat Commun. 2023;14:5417.
Sanders LM, Chandra R, Zebarjadi N, Beale HC, Lyle AG, Rodriguez A, et al. Machine learning multi-omics analysis reveals cancer driver dysregulation in pan-cancer cell lines compared to primary tumors. Commun Biol. 2022;5:1367.
Peng D, Gleyzer R, Tai WH, Kumar P, Bian Q, Isaacs B, et al. Evaluating the transcriptional fidelity of cancer models. Genome Med. 2021;13:73.
Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11–32.
Jan YH, Tan KT, Chen SJ, Yip TTC, Lu CT, Lam AK. Comprehensive assessment of actionable genomic alterations in primary colorectal carcinoma using targeted next-generation sequencing. Br J Cancer. 2022;127:1304–11.
Tabernero J, Ros J, Elez E. The Evolving Treatment Landscape in BRAF-V600E-Mutated Metastatic Colorectal Cancer. Am Soc Clin Oncol Educ Book. 2022;42:1–10.
Strickler JH, Cercek A, Siena S, Andre T, Ng K, Van Cutsem E, et al. Tucatinib plus trastuzumab for chemotherapy-refractory, HER2-positive, RAS wild-type unresectable or metastatic colorectal cancer (MOUNTAINEER): a multicentre, open-label, phase 2 study. Lancet Oncol. 2023;24:496–508.
Venturini J, Massaro G, Lavacchi D, Rossini D, Pillozzi S, Caliman E, et al. The emerging HER2 landscape in colorectal cancer: the key to unveil the future treatment algorithm?. Crit Rev Oncol Hematol. 2024;204:104515.
Chun YS, Passot G, Yamashita S, Nusrat M, Katsonis P, Loree JM, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg. 2019;269:917–23.
Lillemoe HA, Passot G, Kawaguchi Y, DeBellis M, Glehen O, Chun YS, et al. RAS/TP53 co-mutation is associated with worse survival after concurrent resection of colorectal liver metastases and extrahepatic disease. Ann Surg. 2022;276:357–62.
Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget. 2017;8:112103–15.
Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207–16.
Nebot-Bral L, Brandao D, Verlingue L, Rouleau E, Caron O, Despras E, et al. Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer. 2017;84:290–303.
Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3.
Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discov. 2020;10:1129–39.
Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002.
Mariella E, Grasso G, Miotto M, Buzo K, Reilly NM, Andrei P, et al. Transcriptome-wide gene expression outlier analysis pinpoints therapeutic vulnerabilities in colorectal cancer. Mol Oncol. 2024;18:1460–85.
Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71.
Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–47.
Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16:116.
Arena S, Corti G, Durinikova E, Montone M, Reilly NM, Russo M, et al. A Subset of Colorectal Cancers with Cross-Sensitivity to Olaparib and Oxaliplatin. Clin Cancer Res. 2020;26:1372–84.
Durinikova E, Reilly NM, Buzo K, Mariella E, Chila R, Lorenzato A, et al. Targeting the DNA Damage Response Pathways and Replication Stress in Colorectal Cancer. Clin Cancer Res. 2022;28:3874–89.
Quinn LA, Moore GE, Morgan RT, Woods LK. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979;39:4914–24.
Gock M, Mullins CS, Bergner C, Prall F, Ramer R, Goder A, et al. Establishment, functional and genetic characterization of three novel patient-derived rectal cancer cell lines. World J Gastroenterol. 2018;24:4880–92.
Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, et al. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710–8.
de Bruine AP, Dinjens WN, Pijls MM, vd Linden EP, Rousch MJ, Moerkerk PT, et al. NCI-H716 cells as a model for endocrine differentiation in colorectal cancer. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;62:311–20.
Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA. 2006;103:976–81.
Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet. 2022;54:963–75.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Mertz TM, Baranovskiy AG, Wang J, Tahirov TH, Shcherbakova PV. Nucleotide selectivity defect and mutator phenotype conferred by a colon cancer-associated DNA polymerase delta mutation in human cells. Oncogene. 2017;36:4427–33.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7.
Rowan A, Halford S, Gaasenbeek M, Kemp Z, Sieber O, Volikos E, et al. Refining molecular analysis in the pathways of colorectal carcinogenesis. Clin Gastroenterol Hepatol. 2005;3:1115–23.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507.
Liu Y, Chen H, Bao H, Zhang J, Wu R, Zhu L. Comprehensive characterization of FBXW7 mutational and clinicopathological profiles in human colorectal cancers. Front Oncol. 2023;13:1154432.
Wen L, Chen Z, Ji X, Fong WP, Shao Q, Ren C, et al. Pathological complete response to immune checkpoint inhibitor in patients with colorectal cancer liver metastases harboring POLE exonuclease domain mutation. J Immunother Cancer. 2022;10:e004487.
Li X, Sun K, Liao X, Gao H, Zhu H, Xu R. Colorectal carcinomas with mucinous differentiation are associated with high frequent mutation of KRAS or BRAF mutations, irrespective of quantity of mucinous component. BMC Cancer. 2020;20:400.
Mehrvarz Sarshekeh A, Alshenaifi J, Roszik J, Manyam GC, Advani SM, Katkhuda R, et al. ARID1A Mutation May Define an Immunologically Active Subgroup in Patients with Microsatellite Stable Colorectal Cancer. Clin Cancer Res. 2021;27:1663–70.
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93.
Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45.
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncology. 2017;1:1–16.
van der Meer D, Barthorpe S, Yang W, Lightfoot H, Hall C, Gilbert J, et al. Cell Model Passports-a hub for clinical, genetic and functional datasets of preclinical cancer models. Nucleic Acids Res. 2019;47:D923–D9.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–33.
Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet. 2002;11:1549–60.
Fennell LJ, Kane A, Liu C, McKeone D, Fernando W, Su C, et al. APC mutation marks an aggressive subtype of BRAF mutant colorectal cancers. Cancers. 2020;12:1171.
Schell MJ, Yang M, Teer JK, Lo FY, Madan A, Coppola D, et al. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun. 2016;7:11743.
Kaiser C, Meurice N, Gonzales IM, Arora S, Beaudry C, Bisanz KM, et al. Chemogenomic analysis identifies Macbecin II as a compound specific for SMAD4-negative colon cancer cells. Chem Biol Drug Des. 2010;75:360–8.
Woodford-Richens KL, Rowan AJ, Gorman P, Halford S, Bicknell DC, Wasan HS, et al. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci USA. 2001;98:9719–23.
Frey P, Devisme A, Rose K, Schrempp M, Freihen V, Andrieux G, et al. SMAD4 mutations do not preclude epithelial-mesenchymal transition in colorectal cancer. Oncogene. 2022;41:824–37.
Boot A, van Eendenburg J, Crobach S, Ruano D, Speetjens F, Calame J, et al. Characterization of novel low passage primary and metastatic colorectal cancer cell lines. Oncotarget. 2016;7:14499–509.
Boot A, Oosting J, van Eendenburg JDH, Kuppen PJK, Morreau H, van Wezel T. Methylation associated transcriptional repression of ELOVL5 in novel colorectal cancer cell lines. PLoS One. 2017;12:e0184900.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–76.
Ried T, Meijer GA, Harrison DJ, Grech G, Franch-Exposito S, Briffa R, et al. The landscape of genomic copy number alterations in colorectal cancer and their consequences on gene expression levels and disease outcome. Mol Asp Med. 2019;69:48–61.
Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–95.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D7.
Sondka Z, Dhir NB, Carvalho-Silva D, Jupe S, Madhumita, McLaren K, et al. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 2024;52:D1210–D7.
Sveen A, Johannessen B, Eilertsen IA, Rosok BI, Gulla M, Eide PW, et al. The expressed mutational landscape of microsatellite stable colorectal cancers. Genome Med. 2021;13:142.
Prasad CP, Tripathi SC, Kumar M, Mohapatra P. Passage number of cancer cell lines: Importance, intricacies, and way-forward. Biotechnol Bioeng. 2023;120:2049–55.
Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–62.
Robin T, Capes-Davis A, Bairoch A. CLASTR: The Cellosaurus STR similarity search tool - A precious help for cell line authentication. Int J Cancer. 2020;146:1299–306.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Sveen A, Johannessen B, Tengs T, Danielsen SA, Eilertsen IA, Lind GE, et al. Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med. 2017;9:46.
Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33:125–36.
Leon-Castillo A, Britton H, McConechy MK, McAlpine JN, Nout R, Kommoss S, et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol. 2020;250:323–35.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Acknowledgements
Co-115, COLO 320, EB, FRI, HT-29, Isreco-1, Isreco-3, LS1034, LS174T, TC71, SW480, and VACO 9P cells were kindly provided by Dr. Richard Hamelin (National Institute for Health and Medical Research (INSERM), France). The study was funded by grants from the South-Eastern Norway Regional Health Authority (project number 2023101 to A.S. and project numbers 2024108; 2021058 to R.A.L.), the Norwegian Cancer Society (project number 208336 to A.S. and project number 223319-2021 to R.A.L.), and the Research Council of Norway (project number 287899 to A.S.).
Author information
Authors and Affiliations
Contributions
KCGB, AS, and RAL designed the gene panel. BN, ME, IAE, and MH performed the experiments. CK, IAE, LN, GEL, RIS, RAL, and AS analyzed/interpreted the STR and mutational data. CK, IAE, and AS wrote the manuscript. SHM extracted data on mutations’ pathogenicity from public databases. AS and RAL provided supervision, conceptualizationn and acquired funding. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kranjec, C., Eilertsen, I.A., Nunes, L. et al. Common gene mutations in 103 authenticated colorectal cancer cell lines. Oncogenesis (2026). https://doi.org/10.1038/s41389-026-00599-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-026-00599-0