Abstract
Background
Biallelic deleterious variants in RTTN, which encodes rotatin, are associated with primary microcephaly, polymicrogyria, seizures, intellectual disability, and primordial dwarfism in human infants.
Methods and results
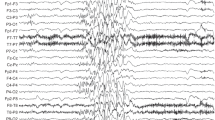
We performed exome sequencing of an infant with primary microcephaly, pontocerebellar hypoplasia, and intractable seizures and his healthy, unrelated parents. We cultured the infant’s fibroblasts to determine primary ciliary phenotype.
Results
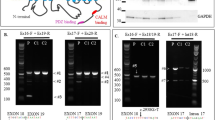
We identified biallelic variants in RTTN in the affected infant: a novel missense variant and a rare, intronic variant that results in aberrant transcript splicing. Cultured fibroblasts from the infant demonstrated reduced length and number of primary cilia.
Conclusion
Biallelic variants in RTTN cause primary microcephaly in infants. Functional characterization of primary cilia length and number can be used to determine pathogenicity of RTTN variants.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mochida, G. H. & Walsh, C. A. Molecular genetics of human microcephaly. Curr. Opin. Neurol. 14, 151–156 (2001).
Adachi, Y. et al. Congenital microcephaly with a simplified gyral pattern: associated findings and their significance. AJNR Am. J. Neuroradiol. 32, 1123–1129 (2011).
Zaqout, S., Morris-Rosendahl, D. & Kaindl, A. M. Autosomal recessive primary microcephaly (MCPH): an update. Neuropediatrics 48, 135–142 (2017).
Desir, J., Cassart, M., David, P., Van Bogaert, P. & Abramowicz, M. Primary microcephaly with ASPM mutation shows simplified cortical gyration with antero-posterior gradient pre- and post-natally. Am. J. Med. Genet. A 146A, 1439–1443 (2008).
Bhat, V. et al. Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clin. Genet. 80, 532–540 (2011).
Rump, P. et al. Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med. Genomics 9, 7 (2016).
Shamseldin, H. et al. RTTN mutations cause primary microcephaly and primordial dwarfism in humans. Am. J. Hum. Genet. 97, 862–868 (2015).
Kheradmand Kia, S. et al. RTTN mutations link primary cilia function to organization of the human cerebral cortex. Am. J. Hum. Genet. 91, 533–540 (2012).
Faisst, A. M., Alvarez-Bolado, G., Treichel, D. & Gruss, P. Rotatin is a novel gene required for axial rotation and left-right specification in mouse embryos. Mech. Dev. 113, 15–28 (2002).
Chatterjee, B., Richards, K., Bucan, M. & Lo, C. Nt mutation causing laterality defects associated with deletion of rotatin. Mamm. Genome 18, 310–315 (2007).
Grandone, A. et al. Expanding the phenotype of RTTN variations: a new family with primary microcephaly, severe growth failure, brain malformations and dermatitis. Clin. Genet. 90, 445–450 (2016).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 (2001).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362 (2014).
Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6, e1001025 (2010).
Cooper, G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005).
Liu, X., Wu, C., Li, C. & Boerwinkle, E. dbNSFPv3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 37, 235–241 (2016).
Rogan, P. K., Faux, B. M. & Schneider, T. D. Information analysis of human splice site mutations. Hum. Mutat. 12, 153–171 (1998).
Reynolds, J. J. et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat. Genet. 49, 537–549 (2017).
Stevens, N. R., Dobbelaere, J., Wainman, A., Gergely, F. & Raff, J. W. Ana3 is a conserved protein required for the structural integrity of centrioles and basal bodies. J. Cell Biol. 187, 355–363 (2009).
Acknowledgements
The authors thank the Exome Aggregation Consortium Database; a full list of contributing groups can be found at http://exac.broadinstitute.org/about. The authors thank the Genotype-Tissue Expression (GTEx) Project, which was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 1 May 2018. This work was supported by grants from the National Institutes of Health (K08 HL105891 (J.A.W.), K12 HL120002 (F.S.C.), R21/33 HL120760 (F.S.C.)), R01 HL128370 (M.R.M.), the Eunice Kennedy Shriver National Institute of Child Health & Human Development (U54 HD087011 (J.S.S.), the Children’s Discovery Institute (F.S.C., J.A.W., M.R.M.), and the Saigh Foundation (F.S.C.).
Author information
Authors and Affiliations
Contributions
Conception and design: J.A.W., D.J.W., M.S., D.B., and F.S.C. Acquisition, analysis, or interpretation of data: J.A.W., D.J.W., P.Y., M.S., D.B., M.V.A., E.B., J.S.S., D.S., M.R.M., and F.S.C. Drafting and revising the manuscript for important intellectual content: J.A.W., D.J.W., P.Y., M.S., D.B., E.B., J.S.S., D.S., B.P.H., M.V.A., T.F., S.K.D., M.R.M., and F.S.C. All authors have approved the final version and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors have seen and approved the submission and take full responsibility for the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wambach, J.A., Wegner, D.J., Yang, P. et al. Functional characterization of biallelic RTTN variants identified in an infant with microcephaly, simplified gyral pattern, pontocerebellar hypoplasia, and seizures. Pediatr Res 84, 435–441 (2018). https://doi.org/10.1038/s41390-018-0083-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-018-0083-z
This article is cited by
-
Pontocerebellar Hypoplasia: a Pattern Recognition Approach
The Cerebellum (2020)