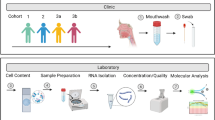
Abstract
Objective
To develop a method to perform multiple tests on a single nasopharyngeal (NP) swab.
Methods
We collected a NP swab on children aged 2–12 years with acute sinusitis and processed it for bacterial culture, viruses, cytokine expression, and 16S ribosomal RNA gene sequencing analysis. During the course of the study, we expand the scope of evaluation to include RNA-sequencing, which we accomplished by cutting the tip of the swab.
Results
Of the 174 children enrolled, 126 (72.4%) had a positive bacterial culture and 121 (69.5%) tested positive for a virus. Cytokine measurement, as judged by adequate levels of a housekeeping enzyme (glyceraldehyde 3-phosphate dehydrogenase), appeared successful. From the samples used for 16S ribosomal sequencing we recovered, on average, 16,000 sequences per sample, accounting for a total of 2646 operational taxonomic units across all samples sequenced. Samples used for RNA-sequencing had a mean RNA integrity number of 6.0. Cutting the tip of the swab did not affect the recovery yield for viruses or bacteria, nor did it affect species richness in microbiome analysis.
Conclusion
We describe a minimally invasive sample collection protocol that allows for multiple diagnostic and research investigations in young children.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Chu, C. Y. et al. The healthy infant nasal transcriptome: a benchmark study. Sci. Rep. 6, 33994 (2016).
de Steenhuijsen Piters, W. A. et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Respir. Crit. Care Med. 194, 1104–1115 (2016).
Chonmaitree, T. et al. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS ONE 12, e0180630 (2017).
Rapola, S., Salo, E., Kiiski, P., Leinonen, M. & Takala, A. K. Comparison of four different sampling methods for detecting pharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae in children. J. Clin. Microbiol. 35, 1077–1079 (1997).
Vuorenoja, K. et al. Detection of Streptococcus pneumoniae carriage by the Binax NOW test with nasal and nasopharyngeal swabs in young children. Eur. J. Clin. Microbiol. Infect. Dis. 31, 703–706 (2012).
Wald, E. R. et al. Clinical practice guideline for the diagnosis and management of acute bacterial sinusitis in children aged 1 to 18 years. Pediatrics 132, e262–e280 (2013).
Daley, P., Castriciano, S., Chernesky, M. & Smieja, M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J. Clin. Microbiol. 44, 2265–2267 (2006).
Smieja, M. et al. Development and evaluation of a flocked nasal midturbinate swab for self-collection in respiratory virus infection diagnostic testing. J. Clin. Microbiol. 48, 3340–3342 (2010).
Leber, A. L. Clinical Microbiology Procedures Handbook 4th edn. (ASM Press, Washington, 2016).
Treanor, J. J. et al. Effectiveness of seasonal influenza vaccines in the United States during a season with circulation of all three vaccine strains. Clin. Infect. Dis. 55, 951–959 (2012).
Talbot, H. K. et al. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J. Med. Virol. 81, 853–856 (2009).
Miller, E. K. et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 123, 98–104 e1 (2009).
Klemenc, J. et al. Real-time reverse transcriptase PCR assay for improved detection of human metapneumovirus. J. Clin. Virol. 54, 371–375 (2012).
Khuri-Bulos, N. et al. Middle East respiratory syndrome coronavirus not detected in children hospitalized with acute respiratory illness in Amman, Jordan, March 2010 to September 2012. Clin. Microbiol. Infect. 20, 678–82 (2014).
Hall, C. B. et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics 132, e341–e348 (2013).
Grijalva, C. G. et al. Concordance between RT-PCR-based detection of respiratory viruses from nasal swabs collected for viral testing and nasopharyngeal swabs collected for bacterial testing. J. Clin. Virol. 60, 309–312 (2014).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Bolotin, S. et al. Development of a novel real-time reverse-transcriptase PCR method for the detection of H275Y positive influenza A H1N1 isolates. J. Virol. Methods 158, 190–194 (2009).
Ali, S. A. et al. Real-world comparison of two molecular methods for detection of respiratory viruses. Virol. J. 8, 332 (2011).
Walters, W. et al. Improved bacterial 16S rRNA gene (V4 and V4–5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, e00009–15 (2016).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Package vegan. https://github.com/vegandevs/vegan. Accessed 16 Apr 2019.
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Poole, A. et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol. 133, 670–8 e12 (2014).
Castro-Nallar, E. et al. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med. Genom. 8, 50 (2015).
Perez-Losada, M., Castro-Nallar, E., Bendall, M. L., Freishtat, R. J. & Crandall, K. A. Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS ONE 10, e0131819 (2015).
Blaschke, A. J. et al. Non-invasive sample collection for respiratory virus testing by multiplex PCR. J. Clin. Virol. 52, 210–214 (2011).
Thwaites, R. S. et al. Nasosorption as a minimally invasive sampling procedure: mucosal viral load and inflammation in primary RSV bronchiolitis. J. Infect. Dis. 215, 1240–1244 (2017).
Chan, K. H., Peiris, J. S., Lim, W., Nicholls, J. M. & Chiu, S. S. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J. Clin. Virol. 42, 65–69 (2008).
Esposito, S. et al. Collection by trained pediatricians or parents of mid-turbinate nasal flocked swabs for the detection of influenza viruses in childhood. Virol. J. 7, 85 (2010).
Acknowledgements
We thank the Microbiology Department at UPMC Children’s Hospital of Pittsburgh. This work was supported by Grant Numbers U01AI118506, 5 UL1 TR001857. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCATS, NIAID, or NIH.
Author information
Authors and Affiliations
Contributions
S.M.C.L., J.M.M., N.S., J.V.W., M.J. and M.K.-L. contributed to conception, design, acquisition of data, analysis and interpretation of data, drafted the article and revised it critically for important intellectual content, and gave final approval of the version to be published. All authors revised it critically for important intellectual content and gave final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lopez, S.M.C., Martin, J.M., Johnson, M. et al. A method of processing nasopharyngeal swabs to enable multiple testing. Pediatr Res 86, 651–654 (2019). https://doi.org/10.1038/s41390-019-0498-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-019-0498-1