Abstract
Background
Paternally expressed gene 10 (PEG10) is believed to be a key imprinted gene involved in placenta formation. However, its role in human folate-related spina bifida (SB) remains unclear.
Methods
The methylation status of the germline differentially methylated region (gDMR) in the PEG10/sarcoglycan epsilon (SGCE) imprinted cluster was compared between SB patients and control samples. Moreover, the influence of ectopic PEG10 expression on apoptosis was assessed to explore the underlying mechanisms related to folate deficiency-induced aberrant gDMR methylation in SB.
Results
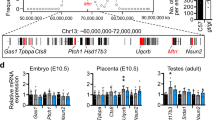
The case group exhibited a significant increase in the methylation level of the gDMR and a marked reduction in the mRNA and protein expression of PEG10 compared with the control group. A prominent negative correlation was found between the folate level in brain tissue and gDMR methylation status (r = −0.62, P = 0.001). A cell model treated with a demethylating agent showed a significant elevation of PEG10 transcription level, as well as other imprinted genes in this cluster. In addition, the inhibition of PEG10 was found to be accompanied by aberrant activation of apoptosis in SB.
Conclusions
Our findings suggest that disturbed gDMR methylation of the PEG10/SGCE cluster due to folate deficiency is involved in SB through aberrant activation of apoptosis.
Impact
-
Disturbed genomic imprinting has been verified to be involved in neural tube defects (NTDs). However, little is known about the effect of ectopic expression of imprinted gene PEG10 on human NTDs.
-
Aberrant methylation status of the germline differentially methylated region (gDMR) of PEG10/SGCE cluster due to folate deficiency has been found to result in the inhibition of PEG10 and has a marked association with an increased occurrence of spina bifida.
-
Inhibited expression of PEG10 partly is found to be related to the abnormal activation of apoptosis in spina bifida.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Avagliano, L. et al. Overview on neural tube defects: from development to physical characteristics. Birth Defects Res. 111, 1455–1467 (2019).
Greene, N. D. & Copp, A. J. Neural tube defects. Annu. Rev. Neurosci. 37, 221–242 (2014).
Czeizel, A. E. & Dudas, I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N. Engl. J. Med. 327, 1832–1835 (1992).
Bartolomei, M. S., Oakey, R. J. & Wutz, A. Genomic imprinting: an epigenetic regulatory system. PLoS Genet. 16, e1008970 (2020).
Perez, J. D., Rubinstein, N. D. & Dulac, C. New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu. Rev. Neurosci. 39, 347–384 (2016).
Abramowitz, L. K. & Bartolomei, M. S. Genomic imprinting: recognition and marking of imprinted loci. Curr. Opin. Genet. Dev. 22, 72–78 (2012).
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R. & Riccio, A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20, 235–248 (2019).
Chang, S. et al. The effect of folic acid deficiency on Mest/Peg1 in neural tube defects. Int. J. Neurosci. 131, 468–477 (2021).
Wang, L. et al. Altered gnas imprinting due to folic acid deficiency contributes to poor embryo development and may lead to neural tube defects. Oncotarget 8, 110797–110810 (2017).
Ono, R. et al. A retrotransposon-derived gene, Peg10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 (2001).
Ono, R. et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106 (2006).
Lu, X. L. et al. Sonic hedgehog signaling affected by promoter hypermethylation induces aberrant Gli2 Expression in spina bifida. Mol. Neurobiol. 53, 5413–5424 (2016).
Xie, T. et al. Peg10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. 18, 112 (2018).
Garcia-Montojo, M., Doucet-O’Hare, T., Henderson, L. & Nath, A. Human endogenous retrovirus-K (Hml-2): a comprehensive review. Crit. Rev. Microbiol. 44, 715–738 (2018).
Peters, J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet 15, 517–530 (2014).
Murphy, S. K., Huang, Z. & Hoyo, C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS ONE 7, e40924 (2012).
Argyraki, M. et al. In-utero stress and mode of conception: impact on regulation of imprinted genes, fetal development and future health. Hum. Reprod. Update 25, 777–801 (2019).
Monk, D. et al. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, Tfpi2/Tfpi2, which requires Ehmt2 and Eed for allelic-silencing. Genome Res. 18, 1270–1281 (2008).
Liu, J. et al. Prevalence and trend of neural tube defects in five counties in Shanxi Province of Northern China, 2000 to 2014. Birth Defects Res. A Clin. Mol. Teratol. 106, 267–274 (2016).
Wu, L. et al. Altered methylation of Igf2 Dmr0 is associated with neural tube defects. Mol. Cell Biochem. 380, 33–42 (2013).
Toschi, P. et al. Maternal peri-conceptional undernourishment perturbs offspring sperm methylome. Reproduction 159, 513–523 (2020).
Wang, L. et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am. J. Clin. Nutr. 91, 1359–1367 (2010).
Edwards, C. A. et al. The evolution of imprinting: chromosomal mapping of orthologues of mammalian imprinted domains in monotreme and marsupial mammals. BMC Evol. Biol. 7, 157 (2007).
Tucci, V., Isles, A. R., Kelsey, G. & Ferguson-Smith, A. C. Genomic imprinting and physiological processes in mammals. Cell 176, 952–965 (2019).
Kelsey, G. & Feil, R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110336 (2013).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017).
Suzuki, S. et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55 (2007).
Peng, Y. P. et al. Peg10 overexpression induced by E2f-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J. Exp. Clin. Cancer Res. 36, 30 (2017).
Wang, C. et al. Peg10 directly regulated by E2fs might have a role in the development of hepatocellular carcinoma. FEBS Lett. 582, 2793–2798 (2008).
Meng, H. et al. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 11, 604–617 (2015).
Okabe, H. et al. Involvement of Peg10 in human hepatocellular carcinogenesis through interaction with Siah1. Cancer Res. 63, 3043–3048 (2003).
Lux, A. et al. Human retroviral Gag- and Gag-Pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1. J. Biol. Chem. 280, 8482–8493 (2005).
Cecconi, F., Piacentini, M. & Fimia, G. M. The involvement of cell death and survival in neural tube defects: a distinct role for apoptosis and autophagy? Cell Death Differ. 15, 1170–1177 (2008).
Acknowledgements
We gratefully acknowledge all the participants in the study. We thank Emma Longworth-Mills, Ph.D., from LiwenBianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
Funding
This project was supported by the National Natural Science Fund of China (81701452, 81670802), the National Natural Science Fund of Beijing (7202018), the National Key Research and Development Program (2016YFC1000502), and the CAMS Initiative for Innovative Medicine (2016-I2M-1-008).
Author information
Authors and Affiliations
Contributions
X.L., T.Z., and L.Wang designed the research; X.L., S.Y., J.M., and P.P. conducted the research; S.W., C.S., L.Wu, and S.C. provided essential reagents or materials; X.L. and S.W. analyzed data or performed statistical analysis; X.L. wrote the manuscript; L.Wang revised the manuscript; X.L. and L.Wang had primary responsibility for the final content, and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
The study protocol was reviewed and approved by the local ethics committee and institute review board of the Capital Institute of Pediatrics, and all participants submitted informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lu, X., Yang, S., Jie, M. et al. Folate deficiency disturbs PEG10 methylation modifications in human spina bifida. Pediatr Res 92, 987–994 (2022). https://doi.org/10.1038/s41390-021-01908-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-021-01908-6