Abstract
Background
Small extracellular vesicles (sEV) play a crucial role in immune responses to viral infection. However, the composition of sEV derived from children with viral pneumonia remains ill defined.
Methods
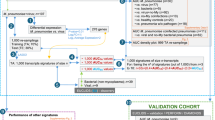
First, we performed mass spectrometry-based label-free proteomic analysis of urinary sEV in 7 children with viral pneumonia, 4 children with Mycoplasma pneumoniae pneumonia and 20 healthy children. Then a total of 33 proteins were selected to validate by multiple reaction monitoring analysis in an independent cohort of 20 healthy children and 29 children with pneumonia.
Results
In the discovery phase, a total of 1621 proteins were identified, while 260 proteins have differential expression in children with viral pneumonia compared to healthy children. Biological pathways primarily associated with neutrophil degranulation, carbohydrate metabolism and endocytosis were enriched in children with viral pneumonia. Finally, the abundance of eight proteins was verified to be significantly higher in children with viral pneumonia than in healthy children.
Conclusions
This pilot study with proteomic profiles of urinary sEV provided insights to the host response to viral pathogen exposure and potential diagnostic biomarkers for children with viral pneumonia, and served as the basis for understanding the fundamental biology of infection.
Impact
-
There were significant differences in the proteomic features of urinary sEV between children with viral pneumonia and those with Mycoplasma pneumoniae pneumonia.
-
Many viral infection-related proteins were identified in urinary sEV and overrepresented in children with viral pneumonia, which facilitates our understanding of the fundamental biology of viral infection.
-
A total of eight proteins (ANPEP, ASAH1, COL11A1, EHD4, HEXB, LGALS3BP, SERPINA1 and SERPING1) were verified as potential biomarkers for the diagnosis of viral pneumonia in children.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The datasets generated during the current study are available in the ProteomeXchange Consortium repository (www.proteomexchange.org) with the dataset identifier PXD036472.
References
Marangu, D. & Zar, H. J. Childhood pneumonia in low-and-middle-income countries: an update. Paediatr. Respir. Rev. 32, 3–9 (2019).
Nascimento-Carvalho, A. C., Ruuskanen, O. & Nascimento-Carvalho, C. M. Comparison of the frequency of bacterial and viral infections among children with community-acquired pneumonia hospitalized across distinct severity categories: a prospective cross-sectional study. BMC Pediatr. 16, 105 (2016).
Bhuiyan, M. U. et al. Role of viral and bacterial pathogens in causing pneumonia among western Australian children: a case-control study protocol. BMJ Open 8, e020646 (2018).
Rodrigues, C. & Groves, H. Community-acquired pneumonia in children: the challenges of microbiological diagnosis. J. Clin. Microbiol. 56, e01318–01317 (2018).
Klompas, M. Does this patient have ventilator-associated pneumonia? JAMA 297, 1583–1593 (2007).
Dela Cruz, C. S. et al. Future research directions in pneumonia. NHLBI Working Group Report. Am. J. Respir. Crit. Care Med. 198, 256–263 (2018).
Thery, C., Zitvogel, L. & Amigorena, S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 (2002).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Raab-Traub, N. & Dittmer, D. P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 15, 559–572 (2017).
Schorey, J. S. & Harding, C. V. Extracellular vesicles and infectious diseases: new complexity to an old story. J. Clin. Invest. 126, 1181–1189 (2016).
Chahar, H. S., Corsello, T., Kudlicki, A. S., Komaravelli, N. & Casola, A. Respiratory syncytial virus infection changes cargo composition of exosome released from airway epithelial cells. Sci. Rep. 8, 387 (2018).
Bedford, J. G. et al. Airway exosomes released during influenza virus infection serve as a key component of the antiviral innate immune response. Front. Immunol. 11, 887 (2020).
Hiemstra, T. F. et al. Human urinary exosomes as innate immune effectors. J. Am. Soc. Nephrol. 25, 2017–2027 (2014).
Yang, L. et al. Lectin microarray combined with mass spectrometry identifies haptoglobin-related protein (HPR) as a potential serologic biomarker for separating nonbacterial pneumonia from bacterial pneumonia in childhood. Proteom. Clin. Appl. 12, e1800030 (2018).
Song, Z. et al. Comprehensive proteomic profiling of urinary exosomes and identification of potential non-invasive early biomarkers of Alzheimer’s disease in 5xfad mouse model. Front. Genet 11, 565479 (2020).
Cheng, J. et al. Proteomic profiling of serum small extracellular vesicles reveals immune signatures of children with pneumonia. Transl. Pediatr. 11, 891–908 (2022).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Wang, S., Kojima, K., Mobley, J. A. & West, A. B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine 45, 351–361 (2019).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Dhondt, B. et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 9, 1736935 (2020).
Kane, M. et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 20, 392–405 (2016).
Wu, T. et al. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics. Data. Innov. (N. Y) 2, 100141 (2021).
Yoshikawa, F. S. Y., Teixeira, F. M. E., Sato, M. N. & Oliveira, L. Delivery of micrornas by extracellular vesicles in viral infections: could the news be packaged? Cells 8, 611 (2019).
Nolte-‘t Hoen, E., Cremer, T., Gallo, R. C. & Margolis, L. B. Extracellular vesicles and viruses: are they close relatives? Proc. Natl Acad. Sci. USA 113, 9155–9161 (2016).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014).
Li, J. et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 14, 793–803 (2013).
Yao, Z. et al. Label-free proteomic analysis of exosomes secreted from THP-1-derived macrophages treated with IFN-alpha identifies antiviral proteins enriched in exosomes. J. Proteome Res. 18, 855–864 (2019).
Sun, L. et al. Exosomes contribute to the transmission of anti-HIV activity from TLR3-activated brain microvascular endothelial cells to macrophages. Antivir. Res. 134, 167–171 (2016).
Crenshaw, B. J., Gu, L., Sims, B. & Matthews, Q. L. Exosome biogenesis and biological function in response to viral infections. Open Virol. J. 12, 134–148 (2018).
Cocozza, F. et al. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 spike protein-containing virus. J. Extracell. Vesicles 10, e12050 (2020).
Xie, F. et al. Engineering extracellular vesicles enriched with palmitoylated ACE2 as COVID-19 therapy. Adv. Mater. 33, e2103471 (2021).
Zhang, J. et al. The interferon-stimulated exosomal HACE2 potently inhibits SARS-CoV-2 replication through competitively blocking the virus entry. Signal Transduct. Target Ther. 6, 189 (2021).
Galani, I. E. & Andreakos, E. Neutrophils in viral infections: current concepts and caveats. J. Leukoc. Biol. 98, 557–564 (2015).
Grudzinska, F. S. et al. Neutrophils in community-acquired pneumonia: parallels in dysfunction at the extremes of age. Thorax 75, 164–171 (2020).
Vargas, A., Roux-Dalvai, F., Droit, A. & Lavoie, J. P. Neutrophil-derived exosomes: a new mechanism contributing to airway smooth muscle remodeling. Am. J. Respir. Cell Mol. Biol. 55, 450–461 (2016).
Genschmer, K. R. et al. Activated PMN exosomes: pathogenic entities causing matrix destruction and disease in the lung. Cell 176, 113–126.e115 (2019).
Yu, Y., Clippinger, A. J. & Alwine, J. C. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 19, 360–367 (2011).
Girdhar, K. et al. Viruses and metabolism: the effects of viral infections and viral insulins on host metabolism. Annu. Rev. Virol. 8, 373–391 (2021).
Vigerust, D. J. & Shepherd, V. L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218 (2007).
Bagdonaite, I. & Wandall, H. H. Global aspects of viral glycosylation. Glycobiology 28, 443–467 (2018).
Watanabe, Y., Bowden, T. A., Wilson, I. A. & Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 1863, 1480–1497 (2019).
Li, Y. et al. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 12, 638573 (2021).
Li, S., Li, S., Wu, S. & Chen, L. Exosomes modulate the viral replication and host immune responses in HBV infection. Biomed. Res. Int. 2019, 2103943 (2019).
Chen, Q. et al. Exosomes mediate horizontal transmission of viral pathogens from insect vectors to plant phloem. Elife 10, e64603 (2021).
Cossart, P. & Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6, a016972 (2014).
Mercer, J., Schelhaas, M. & Helenius, A. Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 (2010).
Ghosh, S. et al. Beta-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183, 1520–1535.e1514 (2020).
Acknowledgements
We are grateful to all the children who took part in the study.
Funding
This study was supported by Grant from the Shanghai Municipal Health Commission (202140111).
Author information
Authors and Affiliations
Contributions
All authors have met the Pediatric Research authorship requirements. L.Y., J.W. and Q.Z. conceived and designed the experiments. J.C., Y.Y., Q.P., J.W. and L.Y. collected clinical samples. J.C., D.J. and S.W. conducted sEV isolation, LC-MS/MS experiments and data analysis. D.J. and Q.Z. drafted the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.J., S.W. and Q.Z. are employees of Wayen Biotechnologies, Inc. The other authors declare that they have no conflicts to declare.
Ethics approval and consent to participate
The study procedure was reviewed and approved by the Ethics Committee of Shanghai Children’s Medical Center. Written informed consent was provided by the patients of the children subjects.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, J., Ji, D., Yin, Y. et al. Proteomic profiling of urinary small extracellular vesicles in children with pneumonia: a pilot study. Pediatr Res 94, 161–171 (2023). https://doi.org/10.1038/s41390-022-02431-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-022-02431-y
This article is cited by
-
Infection-driven proteomic signatures in immune cell–derived extracellular vesicles reflect hemorrhagic stroke outcome
Journal of Neuroinflammation (2025)