Abstract
Background
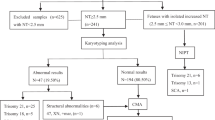
This study aimed to identify large duplications (>5 Mb) that are harmless through long-term clinical follow-ups of fetuses and phenotype analyses of carrier family members.
Methods
We retrospectively analyzed fetuses undergoing prenatal diagnosis and who had >5 Mb chromosomal duplications. Routine karyotyping and single-nucleotide polymorphism array analysis were performed to identify the source and location information of the duplicated segments. Genotype-phenotype analyses were conducted based on genetic information and phenotypes during postnatal follow-up.
Results
Eight eligible cases were included. All fetuses carried maternal or paternal duplications ranging in length from 5.3 to 12.2 Mb. The locations were as follows: 2q32.3q33.1 (Chr2:192322509–199548704), 4q22.1 (Chr4: 88347368–93602855), 4q34.2q35.2 (Chr4:176956406–189189971), 4q34.3q35.2 (Chr4:180613345–189353740), 5p14.3p14.1 (Chr5:19093749–28557664), 10q22.2q23.2 (Chr10:77448435–88786593), 12q21.31q21.32 (Chr12:81983257–87322734), and 13q14.11q14.2 (Chr13: 40825382–47633710). Karyotyping revealed that these duplications occurred within their respective chromosomal regions, except in pedigrees 6 and 7. In the eight pedigrees, the coordinates and lengths of duplicated segments in family members were matched with those in fetuses. Neither the fetuses nor other carriers were clinically symptomatic.
Conclusion
Our findings revealed that the eight pedigrees carrying duplications >5 Mb were asymptomatic, providing new data to inform genetic counseling for the observed segments.
Impact
-
We focused on unrelated fetuses among eight pedigrees who carried duplications of different chromosomal segments. These duplications had been stably transmitted through 2 or 3 generations of normal individuals. Importantly, phenotypic abnormalities were lacking, which was unexpected given that the maximum segment size was approximately 12.2 Mb.
-
We found that duplications in these regions were benign in the context of prenatal genetic counseling. These results provide a foundation for addressing genotype-phenotype correlations. To our knowledge, this is the first description of normal phenotypes in individuals with duplications in these regions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Lupski, J. R. et al. DNA duplication associated with charcot-marie-tooth disease type 1a. Cell 66, 219–232 (1991).
Chance, P. F. et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 72, 143–151 (1993).
Nevado, J. et al. New microdeletion and microduplication syndromes: a comprehensive review. Genet Mol. Biol. 37, 210–219 (2014).
Lee, J. A. & Lupski, J. R. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 52, 103–121 (2006).
Goldenberg, P. An update on common chromosome microdeletion and microduplication syndromes. Pediatr. Ann. 47, e198–e203 (2018).
Martin, C. L., Kirkpatrick, B. E. & Ledbetter, D. H. Copy number variants, aneuploidies, and human disease. Clin. Perinatol. 42, 227–242 (2015).
de Vries, B. B. et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 77, 606–616 (2005).
Tang, M. et al. Duplication of 10q22.3-Q23.3 encompassing Bmpr1a and Ngr3 associated with congenital heart disease, microcephaly, and mild intellectual disability. Am. J. Med. Genet. A 167a, 3174–3179 (2015).
Browne, P. C. et al. Prenatal diagnosis of sub-microscopic partial trisomy 10q using chromosomal microarray analysis in a phenotypically abnormal fetus with normal karyotype. J. Neonatal Perinat. Med. 9, 217–222 (2016).
Hu, H. et al. Prenatal diagnosis and genetic analysis of 21q21.1-Q21.2 aberrations in seven Chinese pedigrees. Front Genet. 12, 731815 (2021).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Riggs, E. R. et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (Clingen). Genet. Med. 22, 245–257 (2020).
Wijedasa, D. Developmental screening in context: adaptation and standardization of the Denver Developmental Screening Test-II (DDST-II) for Sri Lankan children. Child Care Health Dev. 38, 889–899 (2012).
Shaffer, L. G. & Bejjani, B. A. A cytogeneticist’s perspective on genomic microarrays. Hum. Reprod. Update 10, 221–226 (2004).
Callaway, J. L., Shaffer, L. G., Chitty, L. S., Rosenfeld, J. A. & Crolla, J. A. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat. Diagn. 33, 1119–1123 (2013).
Wapner, R. J. et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 367, 2175–2184 (2012).
Levy, B. & Wapner, R. Prenatal diagnosis by chromosomal microarray analysis. Fertil. Steril. 109, 201–212 (2018).
Barber, J. C. Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J. Med. Genet. 42, 609–629 (2005).
Bisgaard, A. M. et al. Transmitted cytogenetic abnormalities in patients with mental retardation: pathogenic or normal variants? Eur. J. Med. Genet. 50, 243–255 (2007).
Collins, R. L. et al. A cross-disorder dosage sensitivity map of the human genome. Cell 185, 3041–3055.e3025 (2022).
Capalbo, A., Rienzi, L. & Ubaldi, F. M. Diagnosis and clinical management of duplications and deletions. Fertil. Steril. 107, 12–18 (2017).
Girirajan, S. et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 367, 1321–1331 (2012).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81971369 to L.J.).
Author information
Authors and Affiliations
Contributions
Huamei Hu designed the study and wrote the article; Ge Huang performed experimental procedures and data analysis; Renke Hou conducted data statistics; Yulin Huang, Yalan Liu, and Xueqian Liao carried out follow-up; Huanhuan Xu conducted chromosome analysis; Juchun Xu performed cell culture; Lupin Jiang proofread the paper and acquired funding; and Dan Wang administered the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no potential conflicts of interest concerning the research, authorship, and/or publication of this article.
Consent to participate
Written informed consent was obtained from the patients.
Ethical approval
This study was approved by the Ethics Committee of Southwest Hospital, Third Military Medical University (Army Medical University). The approval number is (B)KY2021023.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, H., Huang, G., Hou, R. et al. Prenatal diagnosis and genetic analysis: rare familial chromosomal duplications larger than 5 Mb without disease phenotypes. Pediatr Res 97, 2334–2340 (2025). https://doi.org/10.1038/s41390-024-03688-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-024-03688-1