Abstract
Background
The role of Hirschsprung’s disease (HSCR) for the development of inflammatory bowel disease (IBD) and the common pathogenesis of the diseases remains unclear. The objective is to investigate the relationship between HSCR and IBD.
Methods
In our study, the Mendelian randomization approach was employed to analyze the causal relationships. A further search was conducted for differentially expressed genes (DEGs) between disease and control tissues in HSCR and IBD. Subsequently, the potential pathway mechanisms were subjected to an enrichment analysis. Furthermore, the molecular docking was employed to investigate the binding relationship between potential therapeutic targets and drugs.
Results
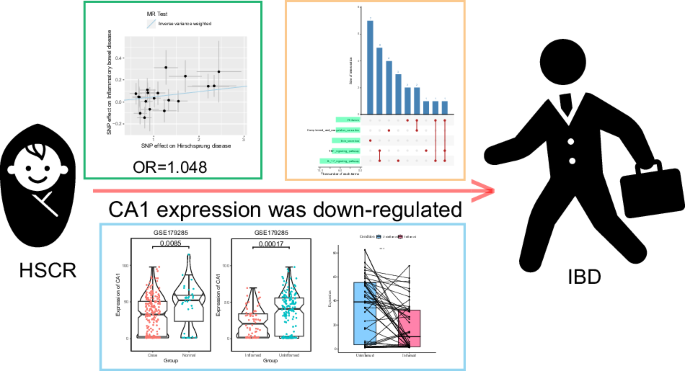
The results show HSCR have an increased risk of developing IBD (IVW: OR = 1.048, P < 0.05; weighted median: OR = 1.065, P < 0.05). A total of 111 DEGs were identified in IBD, while 471 DEGs were observed in HSCR. CA1 was identified as core gene and exhibited lower expression levels in IBD (P < 0.05). Concomitantly, CA1 exhibited reduced expression levels in inflamed tissues. And the TNF and IL17 signaling pathway were found closely related to CA1 expression.
Conclusion
In total, our study shows HSCR promote the occurrence of IBD and reveals pathogenesis. Our results suggest CA1 may provide novel insight for the treatment of HSCR complicated with IBD.
Impact
-
Individuals with HSCR are at a higher risk of developing IBD (IVW: OR = 1.048, P < 0.05; Weighted median: OR = 1.065, P < 0.05).
-
Patients with IBD exhibited lower expression levels of CA1 (P < 0.05). Furthermore, CA1 expression was found to be lower in inflamed tissues (P < 0.05).
-
CA1 may provide novel insight for the treatment of HSCR complicated with IBD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw data used in this study were obtained from publicly available data. Details of the specific data sources are described in the “MATERIALS AND METHODS” section. Relevant results are available in the supplementary material. The codes used in this analysis can be obtained by contacting the corresponding author.
References
Rosen, M. J., Dhawan, A. & Saeed, S. A. Inflammatory Bowel Disease in Children and Adolescents. JAMA Pediatrics 169, 1053–1060 (2015).
Jeong, D. Y. et al. Induction and maintenance treatment of inflammatory bowel disease: A comprehensive review. Autoimmun. Rev. 18, 439–454 (2019).
Bousvaros, A. et al. Differentiating ulcerative colitis from Crohn disease in children and young adults: report of a working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn’s and Colitis Foundation of America. J. Pediatr. Gastroenterol. Nutr. 44, 653–674 (2007).
Khor, B., Gardet, A. & Xavier, R. J. Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011).
Panza, E. et al. Genetics of human enteric neuropathies. Prog. Neurobiol. 96, 176–189 (2012).
Montalva, L. et al. Hirschsprung disease. Nat. Rev. Dis. Prim. 9, 54 (2023).
Lee, H. C. Gene and TET1 association in Hirschsprung disease. Pediatr. Neonatol. 63, 327–328 (2022).
Goldstein, A. M., Thapar, N., Karunaratne, T. B. & De Giorgio, R. Clinical aspects of neurointestinal disease: Pathophysiology, diagnosis, and treatment. Dev. Biol. 417, 217–228 (2016).
Heuckeroth, R. O. Hirschsprung disease - integrating basic science and clinical medicine to improve outcomes. Nat. Rev. Gastroenterol. Hepatol. 15, 152–167 (2018).
Bernstein, C. N. et al. Increased Incidence of Inflammatory Bowel Disease After Hirschsprung Disease: A Population-based Cohort Study. J. Pediatr. 233, 98–104.e102 (2021).
Sutthatarn, P. et al. Hirschsprung-associated inflammatory bowel disease: A multicenter study from the APSA Hirschsprung disease interest group. J. Pediatr. Surg. 58, 856–861 (2023).
Löf Granström, A., Amin, L., Arnell, H. & Wester, T. Increased Risk of Inflammatory Bowel Disease in a Population-based Cohort Study of Patients With Hirschsprung Disease. J. Pediatr. Gastroenterol. Nutr. 66, 398–401 (2018).
Granström, A. L., Ludvigsson, J. F. & Wester, T. Clinical characteristics and validation of diagnosis in individuals with Hirschsprung disease and inflammatory bowel disease. J. Pediatr. Surg. 56, 1799–1802 (2021).
Birney, E. Mendelian Randomization. Cold Spring Harb. Perspect. Med. 12, https://doi.org/10.1101/cshperspect.a041302 (2022).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian Randomization. JAMA 318, 1925–1926 (2017).
Palmer, N. P. et al. Concordance between gene expression in peripheral whole blood and colonic tissue in children with inflammatory bowel disease. PLoS One 14, e0222952 (2019).
Li, K. et al. Molecular Comparison of Adult and Pediatric Ulcerative Colitis Indicates Broad Similarity of Molecular Pathways in Disease Tissue. J. Pediatr. Gastroenterol. Nutr. 67, 45–52 (2018).
Keir, M. E. et al. Regulation and Role of αE Integrin and Gut Homing Integrins in Migration and Retention of Intestinal Lymphocytes during Inflammatory Bowel Disease. J. Immunol. 207, 2245–2254 (2021).
Elsworth, B. et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv, https://doi.org/10.1101/2020.08.10.244293 (2020).
Tang, C. S. et al. Fine mapping of the 9q31 Hirschsprung’s disease locus. Hum. Genet. 127, 675–683 (2010).
Garcia-Etxebarria, K. et al. Local genetic variation of inflammatory bowel disease in Basque population and its effect in risk prediction. Sci. Rep. 12, 3386 (2022).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Bhattacharya, S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 180015 (2018).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–d613 (2019).
Ru, Y. et al. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 42, e133 (2014).
Rauluseviciute, I. et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad1059 (2023).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–d1082 (2018).
Freshour, S. L. et al. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 49, D1144–d1151 (2021).
Zdrazil, B. et al. The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res., https://doi.org/10.1093/nar/gkad1004 (2023).
Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–d1380 (2023).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Pontén, F., Jirström, K. & Uhlen, M. The Human Protein Atlas–a tool for pathology. J. Pathol. 216, 387–393 (2008).
Meng, C. et al. Gut microbiome and risk of ischaemic stroke: a comprehensive Mendelian randomization study. Eur. J. Prevent. Cardiol. 30, 613–620 (2023).
Burgess, S., Scott, R. A., Timpson, N. J., Davey Smith, G. & Thompson, S. G. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552 (2015).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 40, 304–314 (2016).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Soret, R. et al. Glial Cell-Derived Neurotrophic Factor Induces Enteric Neurogenesis and Improves Colon Structure and Function in Mouse Models of Hirschsprung Disease. Gastroenterology 159, 1824–1838.e1817 (2020).
He, E. et al. The potential effects and mechanism of echinacoside powder in the treatment of Hirschsprung’s Disease. Math. Biosci. Eng. 20, 14222–14240 (2023).
Tomuschat, C., O’Donnell, A. M., Coyle, D. & Puri, P. Altered expression of IL36γ and IL36 receptor (IL1RL2) in the colon of patients with Hirschsprung’s disease. Pediatr. Surg. Int. 33, 181–186 (2017).
Tomuschat, C., O’Donnell, A. M., Coyle, D. & Puri, P. Increased Act1/IL-17R expression in Hirschsprung’s disease. Pediatr. Surg. Int. 32, 1201–1207 (2016).
Nakamura, H., Lim, T. & Puri, P. Inflammatory bowel disease in patients with Hirschsprung’s disease: a systematic review and meta-analysis. Pediatr. Surg. Int. 34, 149–154 (2018).
Saez, A., Herrero-Fernandez, B., Gomez-Bris, R., Sánchez-Martinez, H. & Gonzalez-Granado, J. M. Pathophysiology of Inflammatory Bowel Disease: Innate Immune System. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms24021526 (2023).
Verstockt, B. et al. Sphingosine 1-phosphate modulation and immune cell trafficking in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 19, 351–366 (2022).
Higashiyama, M. & Hokaria, R. New and Emerging Treatments for Inflammatory Bowel Disease. Digestion 104, 74–81 (2023).
Skrivankova, V. W. et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA 326, 1614–1621 (2021).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018).
Chen, X. et al. Intestinal proinflammatory macrophages induce a phenotypic switch in interstitial cells of Cajal. J. Clin. Invest. 130, 6443–6456 (2020).
Meir, M. et al. Intestinal Epithelial Barrier Maturation by Enteric Glial Cells Is GDNF-Dependent. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22041887 (2021).
Schumacher, M. A. et al. ErbB4 signaling stimulates pro-inflammatory macrophage apoptosis and limits colonic inflammation. Cell Death Dis. 8, e2622 (2017).
Roberts, R. L. et al. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 9, 561–565 (2008).
Rioux, J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Neurath, M. F. Targeting cytokines in inflammatory bowel disease. Sci. Transl. Med. 14, eabq4473 (2022).
Boland, B. S. et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 5, https://doi.org/10.1126/sciimmunol.abb4432 (2020).
Geremia, A., Biancheri, P., Allan, P., Corazza, G. R. & Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 13, 3–10 (2014).
Boros, É. et al. Elevated Expression of AXL May Contribute to the Epithelial-to-Mesenchymal Transition in Inflammatory Bowel Disease Patients. Mediators Inflamm. 2018, 3241406 (2018).
Hang, S. et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 576, 143–148 (2019).
Tange, K. et al. Oral administration of human carbonic anhydrase I suppresses colitis in a murine inflammatory bowel disease model. Sci. Rep. 12, 17983 (2022).
Minichová, L., Škultéty, Ľ. & Lakota, J. Autoimmune phenomena and spontaneous tumour regression. The role of carbonic anhydrase I. J. Cell. Mol. Med. 25, 5339–5340 (2021).
Schmitt, H., Neurath, M. F. & Atreya, R. Role of the IL23/IL17 Pathway in Crohn’s Disease. Front. Immunol. 12, 622934 (2021).
Cui, G., Fan, Q., Li, Z., Goll, R. & Florholmen, J. Evaluation of anti-TNF therapeutic response in patients with inflammatory bowel disease: Current and novel biomarkers. EBioMedicine 66, 103329 (2021).
Acknowledgements
We would like to acknowledge the public databases including GEO, IEU, GO, KEGG, GEPIA, STRING, JASPAR, Cmap, Drugbank, DGIdb, ChEMBL and HPA for their contributions to human medicine in which they share vast volumes of data. We are also grateful to the majority of researchers who are willing to publicly upload their experimental data. Thanks to the authors of the R package for their contribution to the advancement of Bioinformatics.
Funding
This work was supported in part by Tianjin Municipal Education Commission Research Plan Project (Natural Science) (2023YXZD12) and Tianjin University “Medicine+” Special Fund, Tianjin Natural Science Foundation (22JCZDJC00230).
Author information
Authors and Affiliations
Contributions
Research idea and design: Enyang He, Xiaohong Die and Hualei Cui. Data acquisition: Enyang He, Hailan Zhao and WeiZhao. Data analysis: Enyang He, Bowen Shi, Wenjin Sun, Miao Jia and Kaili Chang. Manuscript Writing: Enyang He,WeiFeng, Bowen Shi and Hongyv Jiang. Reviewing: Enyang He, Liang Dong and Hualei Cui.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was based on open-source data from multiple databases. Ethical approval has been provided for the patients involved in these databases. Therefore, there are no ethical issues with this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, E., Shi, B., Jia, M. et al. Hirschsprung’s disease may increase the incidence of inflammatory bowel disease through alterations in CA1. Pediatr Res 98, 1580–1590 (2025). https://doi.org/10.1038/s41390-025-03938-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-025-03938-w