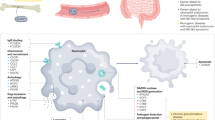
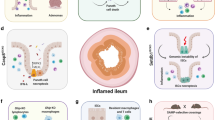
Abstract
Background
Pediatric Crohn’s disease (CD) is a chronic inflammatory bowel disorder that poses significant health risks to children. Although the precise etiology of CD remains elusive, further exploration is needed to identify diagnostic biomarkers and therapeutic targets.
Methods
This study utilized single-cell and bulk RNA sequencing data derived from ileal and colonic biopsy samples to explore the molecular mechanisms and cell types associated with CD, as well as to pinpoint potential biomarkers and therapeutic targets.
Results
The results revealed a more pronounced alteration in both the quantity and functional state of neutrophils in the CD cohort compared to those with ulcerative colitis and healthy controls. Neutrophils were present in higher proportions in the CD group, primarily in an activated state, potentially correlating with the presence of deep ulcerations and inflammatory histopathological features. Additionally, neutrophil interactions with other cell types were markedly enhanced in the CD group, making neutrophils the dominant participants in cell-to-cell communications. Further analysis indicated a shift in neutrophil phenotype from pro-inflammatory and antimicrobial to tissue-repairing, which may contribute to the progression and exacerbation of CD.
Conclusion
IL1B, ICAM1, CXCL1, and CXCL9, primarily expressed in neutrophils, were potential biomarkers for CD. Neutrophils might be considered a potential target for pediatric CD.
Impact Statement
-
This study demonstrated that patients with CD exhibited a greater proportion of activated neutrophils, with enhanced interactions between neutrophils and all other cell types, resulting in neutrophils contributing the most cell-cell interactions within the CD gut.
-
Neutrophils in the CD gut transition from a pro-inflammatory and antibacterial phenotype to one that promotes tissue healing, potentially influencing the progression and exacerbation of CD. Neutrophils represent a promising therapeutic target in pediatric CD.
-
Hub genes associated with CD, including IL1B, ICAM1, CXCL1, and CXCL9, are predominantly expressed in neutrophils, positioning them as promising diagnostic biomarkers for CD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data supporting our research can be obtained from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The data generated during the current study are available from the corresponding author on reasonable request.
References
Rosen, M. J., Dhawan, A. & Saeed, S. A. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 169, 1053–1060 (2015).
Ye, Y., Manne, S., Treem, W. R. & Bennett, D. Prevalence of inflammatory bowel disease in pediatric and adult populations: recent estimates from large national databases in the United States, 2007–2016. Inflamm. Bowel Dis. 26, 619–625 (2019).
Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778 (2017).
Benchimol, E. I. et al. Trends in epidemiology of pediatric inflammatory bowel disease in Canada: distributed network analysis of multiple population-based provincial health administrative databases. Am. J. Gastroenterol. 112, 1120–1134 (2017).
Dolinger, M., Torres, J. & Vermeire, S. Crohn’s disease. Lancet 403, 1177–1191 (2024).
Pigneur, B. et al. Natural history of Crohn’s disease: comparison between childhood- and adult-onset disease. Inflamm. Bowel Dis. 16, 953–961 (2010).
Mitchel, E. B. & Rosh, J. R. Pediatric management of Crohn’s disease. Gastroenterol. Clin. North Am. 51, 401–424 (2022).
Agrawal, M., Spencer, E. A., Colombel, J. F. & Ungaro, R. C. Approach to the management of recently diagnosed inflammatory bowel disease patients: a user’s guide for adult and pediatric gastroenterologists. Gastroenterology 161, 47–65 (2021).
Yu, Y. R. & Rodriguez, J. R. Clinical presentation of Crohn’s, ulcerative colitis, and indeterminate colitis: symptoms, extraintestinal manifestations, and disease phenotypes. Semin. Pediatr. Surg. 26, 349–355 (2017).
Laass, M. W., Roggenbuck, D. & Conrad, K. Diagnosis and classification of Crohn’s disease. Autoimmun. Rev. 13, 467–471 (2014).
Roda, G. et al. Crohn’s disease. Nat. Rev. Dis. Prim. 6, 22 (2020).
Xuan, J., Yu, Y., Qing, T., Guo, L. & Shi, L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 340, 284–295 (2013).
Hong, M. et al. Rna sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 13, 166 (2020).
Stark, R., Grzelak, M. & Hadfield, J. Rna sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656 (2019).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Langfelder, P. & Horvath, S. Wgcna: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005).
Xu, L., Xiao, T., Xu, L. & Yao, W. Identification of therapeutic targets and prognostic biomarkers in cholangiocarcinoma via Wgcna. Front. Oncol. 12, 977992 (2022).
Szklarczyk, D. et al. String V11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Nick, T. G. & Hardin, J. M. Regression modeling strategies: an illustrative case study from medical rehabilitation outcomes research. Am. J. Occup. Ther. 53, 459–470 (1999).
Vickers, A. J. & Elkin, E. B. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Mak. 26, 565–574 (2006).
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling tumor infiltrating immune cells with cibersort. Methods Mol. Biol. 1711, 243–259 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Jin, S. et al. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 12, 1088 (2021).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Hanzelmann, S., Castelo, R. & Guinney, J. Gsva: gene set variation analysis for microarray and Rna-Seq data. BMC Bioinforma. 14, 7 (2013).
Hackert, N. S. et al. Human and mouse neutrophils share core transcriptional programs in both homeostatic and inflamed contexts. Nat. Commun. 14, 8133 (2023).
Danne, C., Skerniskyte, J., Marteyn, B. & Sokol, H. Neutrophils: from ibd to the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 21, 184–197 (2024).
Garrido-Trigo, A. et al. Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat. Commun. 14, 4506 (2023).
Torres, J., Mehandru, S., Colombel, J. F. & Peyrin-Biroulet, L. Crohn’s disease. Lancet 389, 1741–1755 (2017).
Feuerstein, J. D. & Cheifetz, A. S. Crohn’s disease: epidemiology, diagnosis, and management. Mayo Clin. Proc. 92, 1088–1103 (2017).
Gomollon, F. et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn’s disease 2016: part 1: diagnosis and medical management. J. Crohns Colitis 11, 3–25 (2017).
Nahon, S. et al. Diagnostic delay in a French cohort of Crohn’s disease patients. J. Crohns Colitis 8, 964–969 (2014).
Maconi, G. et al. The impact of symptoms, irritable bowel syndrome pattern and diagnostic investigations on the diagnostic delay of Crohn’s disease: a prospective study. Dig. Liver Dis. 47, 646–651 (2015).
Vavricka, S. R. et al. Systematic evaluation of risk factors for diagnostic delay in inflammatory bowel disease. Inflamm. Bowel Dis. 18, 496–505 (2012).
Schoepfer, A. M. et al. Diagnostic delay in Crohn’s disease is associated with a complicated disease course and increased operation rate. Am. J. Gastroenterol. 108, 1744–1753 (2013).
Nahon, S. et al. Diagnostic delay is associated with a greater risk of early surgery in a French cohort of Crohn’s disease patients. Dig. Dis. Sci. 61, 3278–3284 (2016).
Magro, F. et al. European consensus on the histopathology of inflammatory bowel disease. J. Crohns Colitis 7, 827–851 (2013).
Lopez, R. N. et al. Fecal biomarkers in inflammatory bowel disease. J. Gastroenterol. Hepatol. 32, 577–582 (2017).
Zollner, A. et al. Faecal biomarkers in inflammatory bowel diseases: calprotectin versus lipocalin-2-a comparative study. J. Crohns Colitis 15, 43–54 (2021).
Mortha, A. et al. Neutralizing anti-granulocyte macrophage-colony stimulating factor autoantibodies recognize post-translational glycosylations on granulocyte macrophage-colony stimulating factor years before diagnosis and predict complicated Crohn’s disease. Gastroenterology 163, 659–670 (2022).
Fernandes, F. P., Leal, V. N. C., Souza de Lima, D., Reis, E. C. & Pontillo, A. Inflammasome genetics and complex diseases: a comprehensive review. Eur. J. Hum. Genet. 28, 1307–1321 (2020).
Filik, L. The neutrophil respiratory burst and bacterial digestion in Crohn’s disease. Dig. Dis. Sci. 56, 1921 (2011).
Yu, H. H., Yang, Y. H. & Chiang, B. L. Chronic granulomatous disease: a comprehensive review. Clin. Rev. Allergy Immunol. 61, 101–113 (2021).
Visser, G. et al. Neutropenia, neutrophil dysfunction, and inflammatory bowel disease in glycogen storage disease type Ib: results of the European study on glycogen storage disease type I. J. Pediatr. 137, 187–191 (2000).
Marks, D. J., Rahman, F. Z., Sewell, G. W. & Segal, A. W. Crohn’s disease: an immune deficiency state. Clin. Rev. Allergy Immunol. 38, 20–31 (2010).
Acknowledgements
The authors sincerely acknowledge the study participants who donated the samples. The study was supported by the Youth Program of the National Natural Science Foundation of China (No. 81902439).
Author information
Authors and Affiliations
Contributions
L.X. and T.X. conceived the idea, designed the study, analyzed data, performed most of the experiments, and drafted the manuscript. L.X. and B.Z. assisted in designing the study, data analysis, drafting the manuscript, and sample collection. Y.W. supervised the entire project.
Corresponding authors
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, L., Xiao, T., Xu, L. et al. Bulk and single-cell RNA sequencing reveal the roles of neutrophils in pediatric Crohn’s disease. Pediatr Res 98, 1950–1959 (2025). https://doi.org/10.1038/s41390-025-03961-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-025-03961-x