Abstract
Background
Bronchopulmonary dysplasia (BPD) is a prevalent respiratory disease in premature infants and is accompanied by impaired lung function, increased infection risk, and other long-term complications. This study aimed to elucidate the molecular mechanisms of BPD, especially mitophagy.
Methods
Bioinformatics analyses were performed to identify differentially expressed genes (DEGs) in BPD. Weighted gene co-expression network analysis (WGCNA) was used to explore gene modules associated with mitophagy, functional enrichment analyses to identify key biological processes, and immune infiltration to assess immune cell differences.
Results
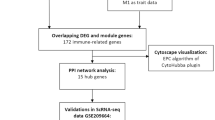
Among the 720 DEGs identified, 419 were upregulated and 301 were downregulated: these may serve as potential BPD biomarkers. WGCNA revealed that the turquoise module was strongly related to mitophagy (r = −0.6061, p < 0.05), indicating its significance in BPD pathogenesis. Enrichment analyses highlighted leukocyte migration and neutrophil extracellular trap formation, suggesting immune-mediated inflammatory response. Eight hub genes (S100P, CDC42EP3, CEACAM3, CKLF, RGL4, DOK3, B4GALT5, and MCEMP1) were identified as potential therapeutic targets. Immune infiltration analysis revealed significant differences in neutrophils and activated CD8+T cells, underscoring the immune system’s role in BPD.
Conclusion
Key molecular players and pathways involved in BPD were elucidated, providing insights for future targeted therapies addressing immunity and mitophagy in BPD.
Impact
-
This study identifies CEACAM3 and CDC42EP3 as key genes involved in mitophagy and immune dysregulation in bronchopulmonary dysplasia (BPD).
-
It provides novel insights into the TNF-α/NF-κB signaling pathway and its role in the pathogenesis of BPD.
-
This study advances biomarker discovery by associating CEACAM3 with neutrophil infiltration and CDC42EP3 with CD8+ T cell activity.
-
The selected machine learning and bioinformatics approaches enhance the diagnostic accuracy and therapeutic targeting of BPD.
-
These findings lay the foundation for future translational research in guiding personalized interventions for high-risk neonates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets utilized here can be obtained in online repositories. The article/supplementary material lists the names of the repository/repositories as well as respective accession numbers.
References
Guimarães, H. et al. Respiratory outcomes and atopy in school-age children who were preterm at birth, with and without bronchopulmonary dysplasia. Clinical 66, 425–430 (2011).
Dankhara, N., Holla, I., Ramarao, S., Kalikkot & Thekkeveedu, R. Bronchopulmonary dysplasia: pathogenesis and pathophysiology. J. Clin. Med. 12, 4207 (2023).
Doyle, L. W. et al. Bronchopulmonary dysplasia in very low birth weight subjects and lung function in late adolescence. Pediatrics 118, 108–113 (2006).
Farhath, S. et al. Pepsin, a marker of gastric contents, is increased in tracheal aspirates from preterm infants who develop bronchopulmonary dysplasia. Pediatrics 121, e253–e259 (2008).
Mohammadi, A., Higazy, R. & Gauda, E. B. PGC-1α activity and mitochondrial dysfunction in preterm infants. Front. Physiol. 13, 997619 (2022).
Shah, D., Das, P. & Bhandari, V. Mitochondrial dysfunction in bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 197, 1363 (2018).
Lu, Y. et al. Cellular mitophagy: mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics 13, 736–766 (2023).
Sharma, A., Ahmad, S., Ahmad, T., Ali, S. & Syed, M. A. Mitochondrial dynamics and mitophagy in lung disorders. Life Sci. 284, 119876 (2021).
De, R. et al. Acute mental stress induces mitochondrial bioenergetic crisis and hyper-fission along with aberrant mitophagy in the gut mucosa in rodent model of stress-related mucosal disease. Free Radic. Biol. Med. 113, 424–438 (2017).
Yu, X. et al. Hyperoxia exposure arrests alveolarization in neonatal rats via PTEN-induced putative kinase 1-Parkin and Nip3-like protein X-mediated mitophagy disorders. Int. J. Mol. Med. 46, 2126–2136 (2020).
Storti, M. et al. Time-resolved transcriptomic profiling of the developing rabbit’s lungs: Impact of premature birth and implications for modelling bronchopulmonary dysplasia. Respir. Res. 24, 80 (2023).
Zak, S. M. et al. Clinical outcomes through two years for infants with bronchopulmonary dysplasia and tracheomalacia. Pediatr. Pulmonol. 60, e27383 (2025).
Cannavò, L. et al. Oxidative stress and respiratory diseases in preterm newborns. Int. J. Mol. Sci. 22, 12504 (2021).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Gene Ontology Consortium Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220 (2010).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Ru, B. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202 (2019).
Ito, K. & Murphy, D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacomet. Syst. Pharmacol. 2, e79 (2013).
Jobe, A. H. & Bancalari, E. Bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 163, 1723–1729 (2001).
Chess, P. R. et al. Pathogenesis of bronchopulmonary dysplasia. Semin. Perinatol. 30, 171–178 (2006).
Gilfillan, M., Bhandari, A. & Bhandari, V. Diagnosis and management of bronchopulmonary dysplasia. BMJ 375, n1974 (2021).
Shrestha, A. K. et al. Lung omics signatures in a bronchopulmonary dysplasia and pulmonary hypertension-like murine model. Am. J. Physiol. Lung Cell. Mol. Physiol. 315, L734–L741 (2018).
Yue, L. & Yao, H. Mitochondrial dysfunction in inflammatory responses and cellular senescence: Pathogenesis and pharmacological targets for chronic lung diseases. Br. J. Pharmacol. 173, 2305–2318 (2016).
Chen, Y. et al. Network pharmacology, molecular docking, and experimental verification to reveal the mitophagy-associated mechanism of Tangshen formula in the treatment of diabetic nephropathy. Diabetes Metab. Syndr. Obes. 17, 739–757 (2024).
Wang, Q. & Liu, C. Mitophagy plays a “double-edged sword” role in the radiosensitivity of cancer cells. J. Cancer Res. Clin. Oncol. 150, 14 (2024).
Song, C., Pan, S., Zhang, J., Li, N. & Geng, Q. Mitophagy: a novel perspective for insighting into cancer and cancer treatment. Cell Prolif. 55, e13327 (2022).
Cui, Y. et al. Protein phosphatase 1 regulatory subunit 15 A (PPP1R15A) promoted the progression of gastric cancer by activating cell autophagy under energy stress. J. Exp. Clin. Cancer Res. 44, 52 (2025).
Clementi, A., Virzì, G. M., Battaglia, G. G. & Ronco, C. Neurohormonal, endocrine, and immune dysregulation and inflammation in cardiorenal syndrome. Cardiorenal Med. 9, 265–273 (2019).
Goob, G., Adrian, J., Cossu, C. & Hauck, C. R. Phagocytosis mediated by the human granulocyte receptor CEACAM3 is limited by the receptor-type protein tyrosine phosphatase PTPRJ. J. Biol. Chem. 298, 102269 (2022).
Kuiper, J. W. P., Gregg, H. L., Schüber, M., Klein, J. & Hauck, C. R. Controlling the cytoskeleton during CEACAM3-mediated phagocytosis. Eur. J. Cell Biol. 103, 151384 (2024).
Klaile, E. et al. Antibody ligation of CEACAM1, CEACAM3, and CEACAM6 differentially enhance the cytokine release of human neutrophils in responses to Candida albicans. Cell. Immunol. 371, 104459 (2022).
Saiz-Gonzalo, G. et al. Regulation of CEACAM family members by IBD-associated triggers in intestinal epithelial cells, their correlation to inflammation and relevance to IBD pathogenesis. Front. Immunol. 12, 655960 (2021).
Speer, C. P. Pulmonary inflammation and bronchopulmonary dysplasia. J. Perinatol. 26, S57–S62 (2006).
Fang, C., Tu, H., Li, R., Bi, D. & Shu, G. Bronchopulmonary dysplasia: analysis and validation of ferroptosis-related diagnostic biomarkers and immune cell infiltration features. Pediatr. Res. 96, 1673–1680 (2024).
Tao, Z. et al. Identification and immunological characterization of endoplasmic reticulum stress-related molecular subtypes in bronchopulmonary dysplasia based on machine learning. Front. Physiol. 13, 1084650 (2023).
Yin, Z. et al. Zebrafish FKBP5 facilitates apoptosis and SVCV propagation by suppressing NF-κB signaling pathway. Fish. Shellfish Immunol. 155, 110021 (2024).
Sun, L. et al. SAL protects endothelial cells from H2O2-induced endothelial dysfunction: regulation of inflammation and autophagy by EZH2. Int. Immunopharmacol. 142, 113060 (2024).
Duan, Q. et al. Narciclasine attenuates LPS-induced acute lung injury in neonatal rats through suppressing inflammation and oxidative stress. Bioengineered 11, 801–810 (2020).
Chen, C. et al. Adipose mesenchymal stem cells-derived exosomes attenuated hyperoxia-induced lung injury in neonatal rats via inhibiting the NF-κB signaling pathway. Pediatr. Pulmonol. 59, 2523–2534 (2024).
Wang, M. Y. et al. Over-activation of iNKT cells aggravate lung injury in bronchopulmonary dysplasia mice. Redox Biol. 77, 103370 (2024).
Su, W. et al. CXCR6 orchestrates brain CD8+ T cell residency and limits mouse Alzheimer’s disease pathology. Nat. Immunol. 24, 1735–1747 (2023).
Verma, A. et al. In silico comparative analysis of LRRK2 interactomes from brain, kidney and lung. Brain Res. 1765, 147503 (2021).
Yuan, S., Bi, X., Shayiti, F., Niu, Y. & Chen, P. Relationship between circulating miRNA-222-3p and miRNA-136-5p and the efficacy of docetaxel chemotherapy in metastatic castration-resistant prostate cancer patients. BMC Urol. 24, 275 (2024).
Zhao, K. et al. Methylation of microRNA-129-5P modulates nucleus pulposus cell autophagy by targeting Beclin-1 in intervertebral disc degeneration. Oncotarget 8, 86264–86276 (2017).
Shen, C., Yan, J., Jiang, L. S. & Dai, L. Y. Autophagy in rat annulus fibrosus cells: Evidence and possible implications. Arthritis Res. 13, R132 (2011).
Vestre, K. et al. Rab6 regulates cell migration and invasion by recruiting Cdc42 and modulating its activity. Cell. Mol. Life Sci. 76, 2593–2614 (2019).
Zhang, Q. et al. TRIM56 acts through the IQGAP1-CDC42 signaling axis to promote glioma cell migration and invasion. Cell Death Dis. 14, 178 (2023).
Saeed, I., La Caze, A., Hollmann, M. W., Shaw, P. N. & Parat, M. O. New insights on tramadol and immunomodulation. Curr. Oncol. Rep. 23, 123 (2021).
Webster, L. et al. Understanding buprenorphine for use in chronic pain: Expert opinion. Pain. Med. 21, 714–723 (2020).
Bokiniec, R., Własienko, P., Borszewska-Kornacka, M. & Szymkiewicz-Dangel, J. Evaluation of left ventricular function in preterm infants with bronchopulmonary dysplasia using various echocardiographic techniques. Echocardiography 34, 567–576 (2017).
Wang, C. & Jiang, Z. D. Brainstem auditory abnormality in extremely premature babies and the impact of neonatal bronchopulmonary dysplasia. Acta Obstet. Gynecol. Scand. 97, 545–551 (2018).
Yoon, B. H. et al. Amniotic fluid cytokines (interleukin-6, tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8) and the risk for the development of bronchopulmonary dysplasia. Am. J. Obstet. Gynecol. 177, 825–830 (1997).
Acknowledgements
We thank all contributors to the GEO database and all the participants who made this research possible.
Funding
Tianjin Health Research Project(TJWJ2022QN089) and The Science & Technolgy Development Fund of Tianjin Education Commission for Higher Education (2024KJ052) supported this work.
Author information
Authors and Affiliations
Contributions
C.L. overall design and writing of the paper; Y.W. participated in the design of the study and performed the statistical analysis; X.W. contributed analysis tools; Y.L. participated in the revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The ethics approval is exempted. This study does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, C., Wang, Y., Wang, X. et al. Investigating mitophagy mechanisms in bronchopulmonary dysplasia through bioinformatics. Pediatr Res (2025). https://doi.org/10.1038/s41390-025-04319-z
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41390-025-04319-z