Abstract
Background
Timely identification of serious bacterial infections in children presenting to emergency departments is critical, especially among non-immunocompromised children, where early symptoms can be nonspecific. Although many children receive empiric antibiotic treatment based on clinical suspicion, true bloodstream infection is relatively uncommon, and unnecessary antibiotics can contribute to adverse effects and antimicrobial resistance.
Methods
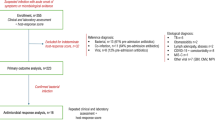
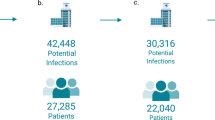
To support individualized decision-making, we developed and evaluated a two-part machine learning framework using retrospective electronic health record data from 5706 pediatric patients aged 3 months to 17 years across six emergency departments. The first model predicted clinical deterioration—defined as admission to intensive care, use of vasopressors, mechanical ventilation, or in-hospital death—among children in whom antibiotics were initially withheld. The second model predicted the likelihood of bacteremia among those who received early empiric antibiotics. Both models were built using XGBoost and evaluated through cross-validation.
Results
Performance was strong, with high area under the curve values and negative predictive values above 96%. Predictive features included supplemental oxygen use, fever, low oxygen saturation, age, and abnormal laboratory values.
Conclusions
This dual-model framework offers interpretable, evidence-based support for early treatment decisions and could improve both patient safety and antibiotic stewardship in pediatric emergency care.
Impact
-
This study introduces a dual machine learning framework that informs early antibiotic decisions in non-immunocompromised pediatric emergency patients.
-
It adds a novel two-model approach: one to predict deterioration when antibiotics are initially withheld, and another to predict bacteremia in those treated.
-
Unlike prior tools, it uses harmonized multi-center EHR data and SHAP-based explain ability to support bedside clinical use.
-
The impact lies in enhancing antibiotic stewardship and patient safety by identifying who may benefit from early antibiotics and who may safely avoid them.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are not publicly available due to institutional data-sharing agreements but may be available from the corresponding author on reasonable request and with appropriate institutional approvals.
References
Hilarius, K. W. E., Skippen, P. W. & Kissoon, N. Early recognition and emergency treatment of sepsis and septic shock in children. Pediatr Emerg Care 36 101–106, https://doi.org/10.1097/PEC.0000000000002043 (2020).
Biban, P. et al. Early recognition and management of septic shock in children. Pediatr Rep 4 e13, https://doi.org/10.4081/pr.2012.e13 (2012).
Dierig, A. et al. Time-to-positivity of blood cultures in children with sepsis. Front Pediatr 6, 222, https://doi.org/10.3389/fped.2018.00222 (2018).
Chiotos, K., D’Arinzo, L., Kitt, E., Ross, R. & Gerber, J. S. Quantifying empiric antibiotic use in US children’s hospitals. Hosp Pediatr 11 e387–e392, https://doi.org/10.1542/hpeds.2021-005950 (2021).
Allen, U. D. Management of infections in the immunocompromised child: general principles. LymphoSign J 3 87–98, https://doi.org/10.14785/lymphosign-2016-0007 (2016).
Bruns, N. & Dohna-Schwake, C. Antibiotics in critically ill children—a narrative review on different aspects of a rational approach. Pediatr Res 91 440–446, https://doi.org/10.1038/s41390-021-01878-9 (2022).
Dar, A., Abram, T. B. & Megged, O. Impact of inadequate empirical antibiotic treatment on outcome of non-critically ill children with bacterial infections. BMC Pediatr 24 324, https://doi.org/10.1186/s12887-024-04793-0 (2024).
Lindell, R. B., Nishisaki, A., Weiss, S. L., Traynor, D. M. & Fitzgerald, J. C. Risk of mortality in immunocompromised children with severe sepsis and septic shock. Crit Care Med 48 1026–1033, https://doi.org/10.1097/CCM.0000000000004329 (2020).
Gerber, J. S., Newland, J., Coffin, S., Hall, M. & Thurm, C. Variability in antibiotic use at children’s hospitals. Pediatrics 126 1067–1073, https://doi.org/10.1542/peds.2010-1275 (2010).
Rostad, C. A., Kanwar, N., Yi, J., Morris, C. & Dien Bard, J. A multicenter evaluation of viral bloodstream detections in children presenting to the emergency department with suspected systemic infection. BMC Pediatr 21, 238, https://doi.org/10.1186/s12887-021-02699-9 (2021).
Zhang, F., Wang, H., Liu, L., Su, T. & Ji, B. Machine learning model for the prediction of gram-positive and gram-negative bacterial bloodstream infection based on routine laboratory parameters. BMC Infect Dis 23 675. https://doi.org/10.1186/s12879-023-08602-4 (2023).
Strich, J. R., Heil, E. L. & Masur, H. Considerations for empiric antimicrobial therapy in sepsis and septic shock in an era of antimicrobial resistance. J Infect Dis 222(Suppl 2), S119–S131, https://doi.org/10.1093/infdis/jiaa221 (2020).
Boerman, A. W. et al. Using machine learning to predict blood culture outcomes in the emergency department: a single-centre, retrospective, observational study. BMJ Open 12 e053332, https://doi.org/10.1136/bmjopen-2021-053332 (2022).
Pang, J. et al. Nonparametric bootstrap methods for interval estimation of the area under the ROC curve with correlated diagnostic test data: application to whole-virus ELISA testing in swine. Front Vet Sci 10, 1274786. https://doi.org/10.3389/fvets.2023.1274786 (2023).
Ponce-Bobadilla, A. V., Schmitt, V., Maier, C. S., Mensing, S. & Stodtmann, S. Practical guide to SHAP analysis: explaining supervised machine learning model predictions in drug development. Clin Transl Sci 17 e70056. https://doi.org/10.1111/cts.70056 (2024).
Mehdary, A., Chehri, A., Jakimi, A. & Saadane, R. Hyperparameter optimization with genetic algorithms and XGBoost: a step forward in smart grid fraud detection. Sensors 24 1230, https://doi.org/10.3390/s24041230 (2024).
Chen T., Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York (NY): ACM; p. 785–794. (2016) https://doi.org/10.1145/2939672.2939785.
Patel, S. J., Chamberlain, D. B. & Chamberlain, J. M. A machine learning approach to predicting need for hospitalization for pediatric asthma exacerbation at the time of emergency department triage. Acad Emerg Med 25 1463–1470, https://doi.org/10.1111/acem.13655 (2018).
Hwang, J. H. et al. Comparison between deep learning and conventional machine learning in classifying iliofemoral deep venous thrombosis upon CT venography. Diagnostics 12 274, https://doi.org/10.3390/diagnostics12020274 (2022).
Hankins, J. D. A little goes a long way: pediatric bloodstream infections and blood culture practices. Clin Microbiol Newsl 44 99–105, https://doi.org/10.1016/j.clinmicnews.2022.06.001 (2022).
Farrell, M. et al. Impact of contaminated blood cultures on children, families, and the health care system. Hosp Pediatr 10 836–843, https://doi.org/10.1542/hpeds.2020-0146 (2020).
Funding
This research was supported by the National Institutes of Health (NIH) through the Small Business Technology Transfer (STTR) program, Award Number 5R41AI167224.
Author information
Authors and Affiliations
Contributions
TV made substantial contributions to the conception and design of the study, led data acquisition and harmonization, developed the machine learning framework, performed model training and evaluation, interpreted results, drafted the article, and approved the final version. IK contributed significantly to study conception and design, provided clinical oversight and multisite coordination, critically revised the manuscript for important intellectual content, and approved the final version. OBM, DH, DCM, HD, MD, RK, and CM contributed to clinical interpretation of data, methodological refinement, validation of modeling strategies, and manuscript writing and revision. MT and FA were involved in data acquisition and contributed to manuscript review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The project was acknowledged by the CNH Institutional Review Board as not constituting human subjects’ research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Velez, T., Badaki-Makun, O., Hirsch, D. et al. Early prediction of antibiotic need and bacteremia risk in non-immunocompromised pediatric emergency patients using machine learning. Pediatr Res (2025). https://doi.org/10.1038/s41390-025-04656-z
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41390-025-04656-z