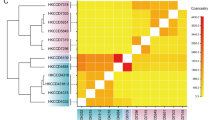
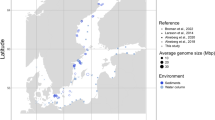
Abstract
An important unanswered question in evolutionary genomics is the source of considerable variation of genomic base composition (GC content) even among organisms that share one habitat. Evolution toward GC-poor genomes has been considered a major adaptive pathway in the oligotrophic ocean, but GC-rich bacteria are also prevalent and highly successful in this environment. We quantify the contribution of multiple factors to the change of genomic GC content of Ruegeria pomeroyi DSS-3, a representative and GC-rich member in the globally abundant Roseobacter clade, using an agent-based model. The model simulates 2 × 108 cells, which allows random genetic drift to act in a realistic manner. Each cell has a whole genome subject to base-substitution mutation and recombination, which affect the carbon and nitrogen requirements of DNA and protein pools. Nonsynonymous changes can be functionally deleterious. Together, these factors affect the growth and fitness. Simulations show that experimentally determined mutation bias toward GC is not sufficient to build the GC-rich genome of DSS-3. While nitrogen availability has been repeatedly hypothesized to drive the evolution of GC content in marine bacterioplankton, our model instead predicts that DSS-3 and its ancestors have been evolving in environments primarily limited by carbon.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Libes S. Introduction to marine biogeochemistry, 2nd ed. Academic Press: Burlington, MA; 2009.
Mizuno CM, Ghai R, Saghaï A, López-García P, Rodriguez-Valera F. Genomes of abundant and widespread viruses from the deep ocean. mBio. 2016;7:e00805–16.
Romero H, Pereira E, Naya H, Musto H. Oxygen and guanine–cytosine profiles in marine environments. J Mol Evol. 2009;69:203–6.
Giovannoni SJ, Thrash JC, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65.
Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110:11463–68.
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359.
Luo H, Huang Y, Stepanauskas R, Tang J. Excess of non-conservative amino acid changes in marine bacterioplankton lineages with reduced genomes. Nat Microbiol. 2017;2:17091.
Luo H, Moran MA. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78:573–87.
Coutinho F, Tschoeke DA, Thompson F, Thompson C. Comparative genomics of Synechococcus and proposal of the new genus Parasynechococcus. PeerJ. 2016;4:e1522.
Sun Y, Powell KE, Sung W, Lynch M, Moran MA, Luo H. Spontaneous mutations of a model heterotrophic marine bacterium. ISME J. 2017;11:1713–18.
Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Ann Rev Mar Sci. 2010;2:305–31.
Long H, Sung W, Kucukyildirim S, Williams E, Miller S, Guo W et al. Evolutionary determinants of genome-wide nucleotide composition. Nature Ecol Evol. 2017;in press.
Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107.
Kelkar YD, Phillips DS, Ochman H. Effects of genic base composition on growth rate in G+C-rich genomes. G3: Genes|Genomes|Genet. 2015;5:1247–52.
Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G + C content in bacterial genes. Proc Natl Acad Sci USA. 2012;109:14504–507.
Lassalle F, Périan S, Bataillon T, Nesme X, Duret L, Daubin V. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 2015;11:e1004941.
Bobay L-M, Ochman H. Impact of recombination on the base composition of bacteria and archaea. Mol Biol Evol. 2017;34:2627–36.
Loewe L, Hill WG. The population genetics of mutations: good, bad and indifferent. Philos Trans R Soc Lond B Biol Sci. 2010;365:1153–67.
Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907.
Hellweger FL, van Sebille E, Fredrick ND. Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science. 2014;345:1346–49.
Kaiser VB, Charlesworth B. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 2009;25:9–12.
Shirani S, Hellweger FL. Neutral evolution and dispersal limitation produce biogeographic patterns in Microcystis aeruginosa populations of lake systems. Microb Ecol. 2017;74:416–26.
Hellweger FL, Clegg RJ, Clark JR, Plugge CM, Kreft J-U. Advancing microbial sciences by individual-based modelling. Nat Rev Micro. 2016;14:461–71.
Dixit PD, Pang TY, Studier FW, Maslov S. Recombinant transfer in the basic genome of Escherichia coli. Proc Natl Acad Sci USA. 2015;112:9070–75.
Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–11.
Scheffer M, Baveco JM, DeAngelis DL, Rose KA, van Nes EH. Super-individuals a simple solution for modelling large populations on an individual basis. Ecol Model. 1995;80:161–70.
Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–13.
Sonnenschein EC, Nielsen KF, D’Alvise P, Porsby CH, Melchiorsen J, Heilmann J, et al. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 2017;11:569–83.
Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041.
Chan L-K, Newton RJ, Sharma S, Smith CB, Rayapati P, Limardo AJ, et al. Transcriptional changes underlying elemental stoichiometry shifts in a marine heterotrophic bacterium. Front Microbiol. 2012;3:159.
Zimmerman AE, Allison SD, Martiny AC. Phylogenetic constraints on elemental stoichiometry and resource allocation in heterotrophic marine bacteria. Environ Microbiol. 2014;16:1398–10.
Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genom Hum Genet. 2010;11:265–89.
Xia X, Xie Z. Protein structure, neighbor effect, and a new index of amino acid dissimilarities. Mol Biol Evol. 2002;19:58–67.
Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. Comparing effective population sizes of dominant marine alphaproteobacteria lineages. Environ Microbiol Rep. 2014;6:167–72.
Dillon MM, Sung W, Sebra R, Lynch M, Cooper VS. Genome-wide biases in the rate and molecular spectrum of spontaneous mutations in Vibrio cholerae and Vibrio fischeri. Mol Biol Evol. 2017;34:93–109.
Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90.
Schluter D, Price T, Mooers AØ, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–11.
Felsenstein J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am J Hum Genet. 1973;25:471–92.
Grzymski JJ, Dussaq AM. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2012;6:71–80.
Sánchez-Baracaldo P, Ridgwell A, Raven John A. A neoproterozoic transition in the marine nitrogen cycle. Curr Biol. 2014;24:652–57.
Luo G, Wang Y, Algeo TJ, Kump LR, Bai X, Yang H, et al Enhanced nitrogen fixation in the immediate aftermath of the latest Permian marine mass extinction. Geology. 2011;39:647–50.
Jenkyns HC. Geochemistry of oceanic anoxic events. Geochem Geophys Geosystems. 2010;11:Q03004.
Zark M, Riebesell U, Dittmar T. Effects of ocean acidification on marine dissolved organic matter are not detectable over the succession of phytoplankton blooms. Sci Adv. 2015;1:e1500531.
Strauss C, Long H, Patterson CE, Te R, Lynch M. Genome-wide mutation rate response to pH change in the coral reef pathogen Vibrio shilonii AK1. mBio. 2017;8:e01021–01017.
Luo H, Csűros M, Hughes AL, Moran MA. Evolution of divergent life history strategies in marine Alphaproteobacteria. mBio. 2013;4:e00373–00313.
Luo H, Thompson LR, Stingl U, Hughes AL. Selection maintains low genomic GC content in marine SAR11 lineages. Mol Biol Evol. 2015;32:2738–48.
Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10.
Sterner RW. On the phosphorus limitation paradigm for lakes. Int Rev Hydrobiol. 2008;93:433–45.
McMahon KD, Read EK. Microbial contributions to phosphorus cycling in eutrophic lakes and wastewater. Annu Rev Microbiol. 2013;67:199–19.
Viklund J, Ettema TJG, Andersson SGE. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol. 2012;29:599–15.
Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–20.
Follows MJ, Dutkiewicz S, Grant S, Chisholm SW. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315:1843–46.
Acknowledgements
We thank Sahar Shirani and Jijun Tang for help with the code development, Rong Zhao for reconstructing the ancestral states of the Roseobacter GC content, Way Sung for providing the Vibrio MA data, and Leong Keat Chan for explaining his microarray data. FLH is supported by the National Science Foundation (1240894 and 1404163); HL is supported by the Hong Kong RGC Early Career Scheme (24101015), the National Natural Science Foundation of China (41576141), the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), and the Direct Grant of the Chinese University of Hong Kong (4930062).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hellweger, F., Huang, Y. & Luo, H. Carbon limitation drives GC content evolution of a marine bacterium in an individual-based genome-scale model. ISME J 12, 1180–1187 (2018). https://doi.org/10.1038/s41396-017-0023-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-017-0023-7
This article is cited by
-
Diversity and distribution of a prevalent Microviridae group across the global oceans
Communications Biology (2024)
-
Metagenome-assembled genomes reveal greatly expanded taxonomic and functional diversification of the abundant marine Roseobacter RCA cluster
Microbiome (2023)
-
Marine viruses disperse bidirectionally along the natural water cycle
Nature Communications (2023)
-
Genome content predicts the carbon catabolic preferences of heterotrophic bacteria
Nature Microbiology (2023)
-
Comparative genomic assessment of members of genus Tenacibaculum: an exploratory study
Molecular Genetics and Genomics (2023)