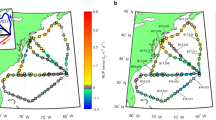
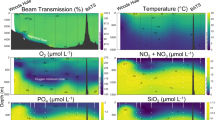
Abstract
Marine net community production (NCP) tracks uptake of carbon by plankton communities and its potential transport to depth. Relationships between marine microbial community composition and NCP currently remain unclear despite their importance for assessing how different taxa impact carbon export. We conducted 16 and 18S rRNA gene (rDNA) sequencing on samples collected across the Western North Atlantic in parallel with high-resolution O2/Ar-derived NCP measurements. Using an internal standard technique to estimate in-situ prokaryotic and eukaryotic rDNA abundances per liter, we employed statistical approaches to relate patterns of microbial diversity to NCP. Taxonomic abundances calculated using internal standards provided valuable context to traditional relative abundance metrics. A bloom in the Mid-Atlantic Bight featured high eukaryote abundances with low eukaryotic diversity and was associated with the harmful algal bloom-forming Aureococcus anophagefferens, phagotrophic algae, heterotrophic flagellates, and particle-associated bacteria. These results show that coastal Aureococcus blooms host a distinct community associated with regionally significant peaks in NCP. Meanwhile, weak relationships between taxonomy and NCP in less-productive waters suggest that productivity across much of this region is not linked to specific microplankton taxa.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–40.
Buesseler KO. The decoupling of production and particulate export in the surface ocean. Glob Biogeochem Cycles. 1998;12:297–310.
Williams R, Follows M. Ocean dynamics and the carbon cycle: principles and mechanisms. Cambridge, United Kingdom: Cambridge University Press; 2011.
Cassar N, Wright SW, Thomson PG, Trull TW, Westwood KJ, de Salas M, et al. The relation of mixed-layer net community production to phytoplankton community composition in the Southern Ocean. Glob Biogeochem Cycles. 2015;29:446–62.
Lin YJ, Cassar N, Marchetti A, Moreno C, Ducklow H, Li ZC. Specific eukaryotic plankton are good predictors of net community production in the Western Antarctic Peninsula. Scientific Reports 2017;7.
Vallina SM, Follows MJ, Dutkiewicz S, Montoya JM, Cermeno P, Loreau M. Global relationship between phytoplankton diversity and productivity in the ocean. Nat Commun. 2014;5:4299.
Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465.
Boyd P, Newton P. Evidence of the potential influence of planktonic community structure on the interannual variability of particulate organic-carbon flux. Deep-Sea Res Part I-Oceanogr Res Pap. 1995;42:619–39.
Richardson TL, Jackson GA. Small phytoplankton and carbon export from the surface ocean. Science. 2007;315:838–840.
Dugdale RC, Goering JJ. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnology Oceanography 1967;12:196.
Li ZC, Cassar N. A mechanistic model of an upper bound on oceanic carbon export as a function of mixed layer depth and temperature. Biogeosciences. 2017;14:5015–27.
Berger WH, Wefer G. Export production: seasonality and intermittency, and paleoceanographic implications. Palaeogeogr, Palaeoclimatol, Palaeoecol. 1990;89:245–54.
Williams, et al. Group report: Export productivity from the photic zone. In: Berger WH, Smetacek V, Wefer G, (eds). Productivity of the ocean: present and past. Hoboken, N.J: John Wiley and Sons; 1989. p. 99–115. PJL
Fawcett SE, Lomas MW, Ward BB, Sigman DM. The counterintuitive effect of summer-to-fall mixed layer deepening on eukaryotic new production in the Sargasso Sea. Glob Biogeochem Cycles. 2014;28:86–102.
Lipschultz F, Bates NR, Carlson CA, Hansell DA. New production in the Sargasso Sea: history and current status. Glob Biogeochem Cycles. 2002;16:1-1–7.
McGillicuddy DJ, Robinson AR, Siegel DA, Jannasch HW, Johnson R, Dickeys T, et al. Influence of mesoscale eddies on new production in the Sargasso Sea. Nature. 1998;394:263–6.
Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep-Sea Res Pt Ii. 2001;48:1405–47.
Lomas MW, Bates NR, Johnson RJ, Knap AH, Steinberg DK, Carlson CA. Two decades and counting: 24-years of sustained open ocean biogeochemical measurements in the Sargasso Sea. Deep-Sea Res Pt Ii. 2013;93:16–32.
Krause JW,Lomas MW,Nelson DM, Biogenic silica at the Bermuda Atlantic Time-series Study site in the Sargasso Sea: Temporal changes and their inferred controls based on a 15-year record. Glob Biogeochem Cycles. 2009;23.
Lomas MW, Steinberg DK, Dickey T, Carlson CA, Nelson NB, Condon RH, et al. Increased ocean carbon export in the Sargasso Sea linked to climate variability is countered by its enhanced mesopelagic attenuation. Biogeosciences. 2010;7:57–70.
Bates NR, Best MHP, Neely K, Garley R, Dickson AG, Johnson RJ. Detecting anthropogenic carbon dioxide uptake and ocean acidification in the North Atlantic Ocean. Biogeosciences. 2012;9:2509–22.
Karl DM, Church MJ. Microbial oceanography and the Hawaii Ocean Time-series programme. Nat Rev Microbiol. 2014;12:699–713.
O’Reilly J, Busch D. Phytoplankton primary production on the northwestern Atlantic shelf. Rapp PV Reun Cons Int Explor Mer. 1984;183:255–68.
Mouw CB, Yoder JA. Primary production calculations in the Mid-Atlantic Bight, including effects of phytoplankton community size structure. Limnol Oceanogr. 2005;50:1232–43.
Bauer JE, Cai W-J, Raymond PA, Bianchi TS, Hopkinson CS, Regnier PAG. The changing carbon cycle of the coastal ocean. Nature. 2013;504:61–70.
Cai W-J. Estuarine and coastal ocean carbon paradox: CO2 sinks or sites of terrestrial carbon incineration? Annu Rev Mar Sci. 2011;3:123–45.
Doney SC. The growing human footprint on coastal and open-ocean biogeochemistry. Science. 2010;328:1512–6.
Lomas MW, Glibert PM, Shiah FK, Smith EM. Microbial processes and temperature in Chesapeake Bay: current relationships and potential impacts of regional warming. Glob Change Biol. 2002;8:51–70.
Brix H, Gruber N, Karl DM, Bates NR. On the relationships between primary, net community, and export production in subtropical gyres. Deep-Sea Res Pt Ii. 2006;53:698–717.
Estapa ML, Siegel DA, Buesseler KO, Stanley RHR, Lomas MW, Nelson NB. Decoupling of net community and export production on submesoscales. Glob Biogeochem Cycles. 2015;29:1266–82.
Mourino-Carballido B, McGillicuddy DJ. Mesoscale variability in the metabolic balance of the Sargasso Sea. Limnol Oceanogr. 2006;51:2675–89.
Treusch AH, Demir-Hilton E, Vergin KL, Worden AZ, Carlson CA, Donatz MG, et al. Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids. Isme J. 2012;6:481–92.
Vergin KL, Done B, Carlson CA, Giovannoni SJ. Spatiotemporal distributions of rare bacterioplankton populations indicate adaptive strategies in the oligotrophic ocean. Aquat Microb Ecol. 2013;71:1–U129.
Cassar N, Barnett B, Bender M, Kaiser J, Hamme R, Tilbrook B. Continuous high-frequency dissolved O-2/Ar measurements by equilibrator inlet mass spectrometry. Anal Chem. 2009;81:1855–64.
Lin Y, Gifford S, Ducklow H, Schofield O, Cassar N (submitted). Towards quantitative marine microbiome community profiling using internal standards.
Smets W, Leff JW, Bradford MA, McCulley RL, Lebeer S, Fierer N. A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol Biochem. 2016;96:145–51.
Moisander PH, Beinart RA, Voss M, Zehr JP. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. Isme J. 2008;2:954–67.
Satinsky BM, Gifford SM, Crump BC, Moran MA. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Microb Metagenomics, Metatranscriptomics, Metaproteomics. 2013;531:237–50.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–22.
Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27:1159–61.
Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31.
Bradley IM, Pinto AJ, Guest JS. Design and evaluation of illumina MiSeq-compatible, 18S rRNA gene-specific primers for improved characterization of mixed phototrophic communities. Appl Environ Microbiol. 2016;82:5878–91.
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6.
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20.
Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: PAired-eND assembler for illumina sequences. BMC Bioinformatics 2012;13.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Schmieder R, Lim YW, Rohwer F, Edwards R. TagCleaner: identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics 2010;11.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96.
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010a;26:266–7.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 2013; 8.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria; 2017
Tang W, Wang S, Batista D, Dehairs F, Gifford S, Gonzalez A et al (submitted). Coastal oceans broaden the biogeography of marine N2 fixation.
Laws EA. Photosynthetic quotients, new production and net community production in the open ocean. Deep-Sea Res Part a-Oceanogr Res Pap. 1991;38:143–67.
Falkowski PG, Flagg CN, Rowe GT, Smith SL, Whitledge TE, Wirick CD. The fate of a spring phytoplankton bloom—export or oxidation. Cont Shelf Res. 1988;8:457–84.
Letscher RT, Moore JK. Modest net autotrophy in the oligotrophic ocean. Glob Biogeochem Cycles. 2017;31:699–708.
Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl Environ Microbiol. 2004;70:4129–35.
Rowe JM, DeBruyn JM, Poorvin L, LeCleir GR, Johnson ZI, Zinser ER, et al. Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. Fems Microbiol Ecol. 2012;79:359–70.
Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. 2013;8:e57923.
Kirchman DL. Processes in microbial ecology. New York: Oxford University Press; 2012.
Rutten TPA, Sandee B, Hofman ART. Phytoplankton monitoring by high performance flow cytometry: a successful approach? Cytom Part A. 2005;64A:16–26.
Gobler CJ, Renaghan MJ, Buck NJ. Impacts of nutrients and grazing mortality on the abundance of Aureococcus anophagefferens during a New York brown tide bloom. Limnol Oceanogr. 2002;47:129–41.
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–10.
Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–40.
Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–32.
Satinsky BM, Fortunato CS, Doherty M, Smith CB, Sharma S, Ward ND et al. Metagenomic and metatranscriptomic inventories of the lower Amazon River, May 2011. Microbiome 2015;3.
Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on pcr amplification of 16s ribosomal-RNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–801.
Barber RT, Hiscock MR. A rising tide lifts all phytoplankton: growth response of other phytoplankton taxa in diatom-dominated blooms. Global Biogeochem Cycles 2006;20.
Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. Isme J. 2011;5:461–72.
Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan L-K, et al. Sizing up metatranscriptomics. ISME J. 2013;7:237–43.
Aitchison J. A new approach to null correlations of proportions. J Int Assoc Math Geol. 1981;13:175–89.
Aitchison J. The statistical-analysis of compositional data. J R Stat Soc Ser B-Methodol. 1982;44:139–77.
Kembel SW, Wu M, Eisen JA, Green JL. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PloS Comput Biol 2012;8.
Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ Microbiol. 2008;10:3349–65.
Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007;9:1233–52.
Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr. 2004;49:784–98.
Figueroa RI, Cuadrado A, Stuken A, Rodriguez F, Fraga S. Ribosomal DNA organization patterns within the dinoflagellate genus alexandrium as revealed by fish: life cycle and evolutionary implications. Protist. 2014;165:343–63.
Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50.
Gobler CJ, Lonsdale DJ, Boyer GL. A review of the causes, effects, and potential management of harmful brown tide blooms caused by Aureococcus anophagefferens (Hargraves et Sieburth). Estuaries. 2005;28:726–49.
Popels LC, Cary SC, Hutchins DA, Forbes R, Pustizzi F, Gobler CJ, et al. The use of quantitative polymerase chain reaction for the detection and enumeration of the harmful alga Aureococcus anophagefferens in environmental samples along the United States East Coast. Limnol Oceanogr-Methods. 2003;1:92–102.
Jones HLJ, Leadbeater BSC, Green JC. Mixotrophy in marine species of chrysochromulina (prymnesiophyceae)– ingestion and digestion of a small green flagellate. J Mar Biol Assoc U K. 1993;73:283–96.
Richardson K. Harmful or exceptional phytoplankton blooms in the marine ecosystem. Adv Mar Biol. 1997;31:301–85.
Graneli E, Edvardsen B, Roelke DL, Hagstrom JA. The ecophysiology and bloom dynamics of Prymnesium spp. Harmful Algae. 2012;14:260–70.
Lage OM, Bondoso J. Planctomycetes and macroalgae, a striking association. Front Microbiol 2014;5.
Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308.
Hatosy SM, Martiny JBH, Sachdeva R, Steele J, Fuhrman JA, Martiny AC. Beta diversity of marine bacteria depends on temporal scale. Ecology. 2013;94:1898–1904.
Cermeño P, Rodríguez-Ramos T, Dornelas M, Figueiras FG, Marañón E, Teixeira IG, et al. Species richness in marine phytoplankton communities is not correlated to ecosystem productivity. Mar Ecol Progress Ser. 2013;488:1–9.
Goebel NL, Edwards CA, Zehr JP, Follows MJ, Morgan SG. Modeled phytoplankton diversity and productivity in the California Current System. Ecol Model. 2013;264:37–47.
Irigoien X, Huisman J, Harris RP. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature. 2004;429:863–7.
Li WKW. Macroecological patterns of phytoplankton in the northwestern North Atlantic Ocean. Nature. 2002;419:154–7.
Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci USA. 2011;108:4352–7.
Caron DA, Alexander H, Allen AE, Archibald JM, Armbrust EV, Bachy C, et al. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat Rev Microbiol. 2017;15:6–20.
Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–98.
Acknowledgements
This research was supported by an NSF-CAREER grant awarded to NC (#1350710) and a Chateaubri and Fellowship awarded to SW. NC was also supported by the "Laboratoire d'Excellence" LabexMER (ANR-10-LABX-19) and co-funded by a grant from the French government under the program "Investissements d'Avenir". RE was supported by an NSF GRFP award (1106401). We thank the staff of the Bermuda Institute of Ocean Sciences as well as the crew and technicians of the R/V Atlantic Explorer for their valuable assistance in organizing and conducting our field study. We are also thankful to Rod Johnson, Bruce Williams, and Natasha McDonald at BIOS for their help with sample shipping and analysis. We are additionally grateful to Karoline Faust for her input on network approaches and to Geoffrey Smith for his help and for our use of his towfish equipment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Subject Category: Geomicrobiology and microbial contributions to geochemical cycles
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, S., Lin, Y., Gifford, S. et al. Linking patterns of net community production and marine microbial community structure in the western North Atlantic. ISME J 12, 2582–2595 (2018). https://doi.org/10.1038/s41396-018-0163-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0163-4