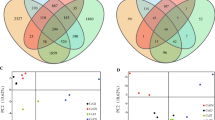
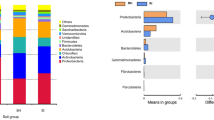
Abstract
Disease-suppressive soils are ecosystems in which plants suffer less from root infections due to the activities of specific microbial consortia. The characteristics of soils suppressive to specific fungal root pathogens are comparable to those of adaptive immunity in animals, as reported by Raaijmakers and Mazzola (Science 352:1392–3, 2016), but the mechanisms and microbial species involved in the soil suppressiveness are largely unknown. Previous taxonomic and metatranscriptome analyses of a soil suppressive to the fungal root pathogen Rhizoctonia solani revealed that members of the Burkholderiaceae family were more abundant and more active in suppressive than in non-suppressive soils. Here, isolation, phylogeny, and soil bioassays revealed a significant disease-suppressive activity for representative isolates of Burkholderia pyrrocinia, Paraburkholderia caledonica, P. graminis, P. hospita, and P. terricola. In vitro antifungal activity was only observed for P. graminis. Comparative genomics and metabolite profiling further showed that the antifungal activity of P. graminis PHS1 was associated with the production of sulfurous volatile compounds encoded by genes not found in the other four genera. Site-directed mutagenesis of two of these genes, encoding a dimethyl sulfoxide reductase and a cysteine desulfurase, resulted in a loss of antifungal activity both in vitro and in situ. These results indicate that specific members of the Burkholderiaceae family contribute to soil suppressiveness via the production of sulfurous volatile compounds.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jousset A, Rochat L, Lanoue A, Bonkowski M, Keel C, Scheu S. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol Plant Microbe Interact. 2010;24:352–8.
Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–63.
Rudrappa T, Czymmek KJ, Paré PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–56.
Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. Microbial populations responsible for specific soil suppressiveness. Annu Rev Phytopathol. 2002;40:309–48.
Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–3.
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100.
Raaijmakers JM, Weller DM. Natural plant protection by 2, 4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact. 1998;11:144–52.
Voort M, Kempenaar M, Driel M, Raaijmakers JM, Mendes R. Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol Lett. 2016;19:375–82.
Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:E1743–52.
de Bruijn I, Cheng X, de Jager V, Expósito RG, Watrous J, Patel N, et al. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genom. 2015;16:1–16.
Gómez Expósito R, Postma J, Raaijmakers JM, de Bruijn I. Diversity and activity of Lysobacter species from disease suppressive soils. Frontiers in Microbiology. 2015; 6:1243.
Gómez Expósito R, de Bruijn I, Postma J, Raaijmakers JM. Current Insights into the Role of Rhizosphere Bacteria in Disease Suppressive Soils. Frontiers in Microbiology. 2017;8:2529.
Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, Van Wezel GP, et al. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol. 2015;6:1081.
Chapelle E, Mendes R, PAHM Bakker, Raaijmakers JM. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10:265–8.
Eberl L, Vandamme P. Members of the genus Burkholderia: Good and bad guys. F1000Res. 2016;5:F1000.
Elliott GN, Chou JH, Chen WM, Bloemberg GV, Bontemps C, Martínez‐Romero E, et al. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N‐limited conditions. Environ Microbiol. 2009;11:762–78.
Carlier AL, Eberl L. The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol. 2012;14:2757–69.
Lewenza S, Sokol PA. Regulation of ornibactin niosynthesis and N-Acyl-l-Homoserine lactone production by cepR in Burkholderia cepacia. J Bacteriol. 2001;183:2212–8.
Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–8.
Partida‐Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45.
Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, et al. Production of the antifungal compound pyrrolnitrin is quorum sensing‐regulated in members of the Burkholderia cepacia complex. Environ Microbiol. 2009;11:1422–37.
Stopnisek N, Zuhlke D, Carlier A, Barberan A, Fierer N, Becher D, et al. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2016;10:253–64.
Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol. 2016;100:5215–29.
Elshafie H, Camele I, Racioppi R, Scrano L, Iacobellis N, Bufo S. In vitro antifungal activity of Burkholderia gladioli pv. agaricicola against some phytopathogenic fungi. Int J Mol Sci. 2012;13:16291.
Groenhagen U, Baumgartner R, Bailly A, Gardiner A, Eberl L, Schulz S, et al. Production of bioactive volatiles by different Burkholderia ambifaria strains. J Chem Ecol. 2013;39:892–906.
Estrada-De Los Santos P, Bustillos-Cristales RO, Caballero-Mellado J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol. 2001;67:2790–8.
Burbage D, Sasser M. A medium selective for Pseudomonas-cepacia. Phytopathology. 1982;72:706.
Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martínez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl Environ Microbiol. 2007;73:5308–19.
Perin L, Martínez-Aguilar L, Castro-González R, Estrada-de los Santos P, Cabellos-Avelar T, Guedes HV, et al. Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl Environ Microbiol. 2006;72:3103–10.
Estrada-de los Santos P, Vinuesa P, Martínez-Aguilar L, Hirsch AM, Caballero-Mellado J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol. 2013;67:51–60.
Payne GW, Ramette A, Rose HL, Weightman AJ, Jones TH, Tiedje JM, et al. Application of a recA gene-based identification approach to the maize rhizosphere reveals novel diversity in Burkholderia species. FEMS Microbiol Lett. 2006;259:126–32.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Letunic I, Bork P. Interactive Tree Of Lifev2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–8.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26.
Garbeva P, Hordijk C, Gerards S, De Boer W. Volatiles produced by the mycophagous soil bacterium Collimonas. FEMS Microbiol Ecol. 2014;87:639–49.
Lommen A, Kools HJ. MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics. 2011;8:719–26.
Tikunov YM, Laptenok S, Hall RD, Bovy A, Vos RCH. MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics. 2011;8:714–8.
Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. Retention index thresholds for compound matching in GC–MS metabolite profiling. J Chromatogr B. 2008;871:182–90.
van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006;7:142.
Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300.
Worley B, Halouska S, Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem. 2013;433:102–4.
Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metab. 2013;1:92–107.
Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. https://doi.org/10.1371/journal.pcbi.1005752.
Core Team R. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2015.
Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA. 2002;99:5890–5.
Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2.
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43.
Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151.
Zumaquero A, Macho AP, Rufián JS, Beuzón CR. Analysis of the role of the type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448a in interaction with the plant. J Bacteriol. 2010;192:4474–88.
van der Voort M, Meijer H, Schmidt Y, Watrous J, Dekkers E, Mendes R, et al. Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front Microbiol. 2015;6:693.
Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, et al. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012;193:997–1008.
Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–12.
Wang C, Wang Z, Qiao X, Li Z, Li F, Chen M, et al. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD−1. FEMS Microbiol Lett. 2013;341:45–51.
Hunziker L, Bönisch D, Groenhagen U, Bailly A, Schulz S, Weisskopf L. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl Environ Microbiol. 2015;81:821–30.
Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–42.
Sawana A, Adeolu M, Gupta RS. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 2014;5:429.
Kreutzer MF, Nett M. Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis. Org Biomol Chem. 2012;10:9338–43.
Hou J, Robbel L, Marahiel Mohamed A. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem Biol. 2011;18:655–64.
Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–31.
Rosconi F, Davyt D, Martínez V, Martínez M, Abin-Carriquiry JA, Zane H, et al. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ Microbiol. 2013;15:916–27.
Kreutzer MF, Kage H, Nett M. Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. J Am Chem Soc. 2012;134:5415–22.
Agnoli K, Lowe CA, Farmer KL, Husnain SI, Thomas MS. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the fur regulon. J Bacteriol. 2006;188:3631–44.
Haq IU, Graupner K, Nazir R, van Elsas JD. The genome of the rungal-interactive soil bacterium Burkholderia terrae BS001—a plethora of outstanding interactive capabilities unveiled. Genome Biol Evol. 2014;6:1652–68.
Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–9.
Wicht DK. The reduced flavin-dependent monooxygenase SfnG converts dimethylsulfone to methanesulfinate. Arch Biochem Biophys. 2016;604:159–66.
De Lorenzo F, Goldberger RF, Steers E, Givol D, Anfinsen CB. Purification and properties of an enzyme from beef liver which catalyzes sulfhydryl-disulfide interchange in proteins. J Biol Chem. 1966;241:1562–7.
Fuchs S, De Lorenzo F, Anfinsen CB. Studies on the mechanism of the enzymic catalysis of disulfide interchange in proteins. J Biol Chem. 1967;242:398–402.
Debussche L, Thibaut D, Cameron B, Crouzet J, Blanche F. Biosynthesis of the corrin macrocycle of coenzyme B12 in Pseudomonas denitrificans. J Bacteriol. 1993;175:7430–40.
McNicholas PM, Chiang RC, Gunsalus RP. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol Microbiol. 1998;27:197–208.
Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23.
Dobbs CG, Hinson WH. A widespread fungistasis in soils. Nature. 1953;172:197–9.
Hora TS, Baker R. Volatile factor in soil fungistasis. Nature. 1970;225:1071–2.
Cheng X, Etalo DW, Mortel JEvd, Dekkers E, Nguyen L, Medema MH, Raaijmakers JM (2017). Genome‐wide analysis of bacterial determinants of plant growth promotion and induced systemic resistance by Pseudomonas fluorescens. Environmental Microbiology 2017;19:4638–4656.
Acknowledgements
J.R. and V.J.C. were supported by a grant from the Netherlands BE-Basic Foundation (project F07.003.01). VC was supported by the Dutch STW-program ‘Back to the Roots’. We thank Elizabeth Yanez Navarrete and Kazuki Fujiguara for their valuable help in the Burkholderia spp. phenotyping and Dr. Florian Rohart for his expert advice in using the R package mixOmics. This is publication number 6497 of the NIOO-KNAW.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Carrión, V.J., Cordovez, V., Tyc, O. et al. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J 12, 2307–2321 (2018). https://doi.org/10.1038/s41396-018-0186-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0186-x
This article is cited by
-
Rapid and sustained differentiation of disease-suppressive phyllosphere microbiomes in tomato following experimental microbiome selection
Environmental Microbiome (2025)
-
Unlocking the potential of ecofriendly guardians for biological control of plant diseases, crop protection and production in sustainable agriculture
3 Biotech (2025)
-
The potential of Paraburkholderia species to enhance crop growth
World Journal of Microbiology and Biotechnology (2025)
-
Microbiome-mediated plant disease resistance: recent advances and future directions
Journal of General Plant Pathology (2025)
-
Mitigating obstacles to monocropping using integrated rhizosphere management
Plant and Soil (2025)