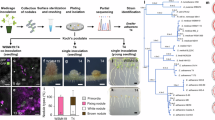
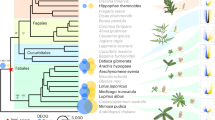
Abstract
Various plant species establish intimate symbioses with bacteria within their aerial organs. The bacteria are contained within nodules or glands often present in distinctive patterns on the leaves in what is commonly referred to as leaf nodule symbiosis. We describe here a highly specific symbiosis between a wild yam species from Madagascar, Dioscorea sansibarensis and bacteria of the species Orrella dioscoreae. Using whole-genome sequencing of plastids and bacteria from wild-collected samples, we show phylogenetic patterns consistent with a dominant vertical mode of transmission of the symbionts. Unique so far among leaf nodule symbioses, the bacteria can be cultured and are amenable to comparative transcriptomics, revealing a potential role in complementing the host’s arsenal of secondary metabolites. We propose a recent establishment of a vertical mode of transmission in this symbiosis which, together with a large effective population size explains the cultivability and apparent lack of genome reductive evolution in O. dioscoreae. We leverage these unique features to reveal pathways and functions under positive selection in these specialized endophytes, highlighting the candidate mechanisms enabling a permanent association in the phyllosphere.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBio. 2015;6:e00621–15.
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–99.
Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10:e1004283.
Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5:5320.
Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, et al. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci USA. 2013;110:E425–34.
Pinto-Carbó M, Eberl L, Gademann K, Carlier A. Leaf nodule symbiosis: function and transmission of obligate bacterial endophytes. Curr Opin Plant Biol. 2018;44:23–31.
Lersten NR, Horner HT Jr. Development and structure of bacterial leaf nodules in Psychotria bacteriophila Val. (Rubiaceae). J Bacteriol. 1967;94:2027–36.
Miller IM. Bacterial leaf nodule symbiosis. Adv Bot Res. 1990;17:163–234.
Carlier A, Fehr L, Pinto-Carbó M, Schäberle T, Reher R, Dessein S, et al. The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the A rdisia crenata leaf nodule symbiosis. Environ Microbiol. 2016;18:2507–22.
Carlier AL, Eberl L. The eroded genome of a leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol. 2012;14:2757–69.
Pinto-Carbó M, Sieber S, Dessein S, Wicker T, Verstraete B, Gademann K, et al. Evidence of horizontal gene transfer between obligate leaf nodule symbionts. ISME J. 2016;10:2092–105.
Sieber S, Carlier A, Neuburger M, Grabenweger G, Eberl L, Gademann K. Isolation and total synthesis of kirkamide, an aminocyclitol from an obligate leaf nodule symbiont. Angew Chem Int Ed. 2015;54:7968–70.
Crüsemann M, Reher R, Schamari I, Brachmann AO, Ohbayashi T, Kuschak M, et al. Heterologous expression, biosynthetic studies, and ecological function of the selective Gq-signaling inhibitor FR900359. Angew Chem Int Ed. 2018;57:836–40.
Carlier A, Cnockaert M, Fehr L, Vandamme P, Eberl L. Draft genome and description of Orrella dioscoreae gen. nov. sp. nov., a new species of Alcaligenaceae isolated from leaf acumens of Dioscorea sansibarensis. Syst Appl Microbiol. 2017;40:11–21.
Miller IM, Reporter M. Bacterial leaf symbiosis in Dioscorea sansibarensis: morphology and ultrastructure of the acuminate leaf glands. Plant Cell Environ. 1987;10:413–24.
Burkill H. The useful plants of west tropical Africa. Volume 1, families A-D. Kew, England: Royal Botanic Gardens, Kew; 1985.
Clark JD, Maaloe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–8.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–95.
Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Li L, Stoeckert CJ, Roos DSC-P. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89.
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 2017;34:2115–22.
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, et al. AntiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41.
Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 2012;7:e34064.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–3.
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M. The metagenomics RAST server-a public resource for the automatic phylo- genetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Zhou W, Nan X, Zheng Z, Wei C, He H. Analysis of inter-individual bacterial variation in gut of Cicada Meimuna mongolica (Hemiptera: Cicadidae). J Insect Sci. 2015;15:131.
Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202.
Russell SL, Cavanaugh CM. Intrahost genetic diversity of bacterial symbionts exhibits evidence of mixed infections and recombinant haplotypes. Mol Biol Evol. 2017;34:2747–61.
Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90.
Fisher RM, Henry LM, Cornwallis CK, Kiers ET, West SA. The evolution of host-symbiont dependence. Nat Commun. 2017;8:15973.
Kuo C-H, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19:1450–4.
Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304.
Mondo SJ, Salvioli A, Bonfante P, Morton JB, Pawlowska TE. Nondegenerative evolution in ancient heritable bacterial endosymbionts of fungi. Mol Biol Evol. 2016;33:2216–31.
Viruel J, Segarra-Moragues JG, Raz L, Forest F, Wilkin P, Sanmartín I, et al. Late Cretaceous-early Eocene origin of yams (Dioscorea, Dioscoreaceae) in the Laurasian Palaearctic and their subsequent Oligocene-Miocene diversification. J Biogeogr. 2016;43:750–62.
Bosdriesz E, Molenaar D, Teusink B, Bruggeman FJ. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 2015;282:2029–44.
Pessi G, Braunwalder R, Grunau A, Omasits U, Ahrens CH, Eberl L. Response of Burkholderia cenocepacia H111 to Micro-Oxia. PLoS ONE 2013;8:e72939.
Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–75.
Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and Role of ryhB in biofilm Formation. Infect Immun. 2005;73:5706–19.
Wu Y, Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191:1248–57.
Lin M-H, Shu J-C, Huang H-Y, Cheng Y-C. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE 2012;7:e34388.
Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102:11076–81.
Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci USA. 2005;102:6502–7.
Helfrich EJN, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316.
Glick BR. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett. 2005;251:1–7.
Kjeldsen KU, Bataillon T, Pinel N, De Mita S, Lund MB, Panitz F, et al. Purifying selection and molecular adaptation in the genome of Verminephrobacter, the heritable symbiotic bacteria of earthworms. Genome Biol Evol. 2012;4:307–15.
Planamente S, Salih O, Manoli E, Albesa‐Jové D, Freemont PS, Filloux A. TssA forms a gp6‐like ring attached to the type VI secretion sheath. EMBO J. 2016;35:1613–27.
Santin YG, Cascales E. Domestication of a housekeeping transglycosylase for assembly of a Type VI secretion system. EMBO Rep. 2017;18:138–49.
Guy B, Krell T, Sanchez V, Kennel A, Manin C, Sodoyer R. Do Th1 or Th2 sequence motifs exist in proteins? Immunol Lett. 2005;96:261–75.
Richter C, Mukherjee O, Ermert D, Singh B, Su Y-C, Agarwal V, et al. Moonlighting of Helicobacter pylori catalase protects against complement-mediated killing by utilising the host molecule vitronectin. Sci Rep. 2016;6:24391.
McCann HC, Nahal H, Thakur S, Guttman DS. Identification of innate immunity elicitors using molecular signatures of natural selection. Proc Natl Acad Sci USA. 2012;109:4215–20.
Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108:10800–7.
Rao AN, Tan AS. Shoot apex and bulbil development in Dioscorea sansibarensis Pax. Bot J Linn Soc. 1976;72:285–98.
Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol. 2010;8:218–30.
Brandvain Y, Goodnight C, Wade MJ. Horizontal transmission rapidly erodes disequilibria between organelle and symbiont genomes. Genetics. 2011;189:397–404.
Douglas AE. Host benefit and the evolution of specialization in symbiosis. Heredity. 1998;81:599–603.
Douglas AE. Conflict, cheats and the persistence of symbioses. New Phytol. 2008;177:849–58.
Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–76.
Bordenstein SR, Paraskevopoulos C, Dunning Hotopp JC, Sapountzis P, Lo N, Bandi C, et al. Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol Biol Evol. 2009;26:231–41.
González-Mula A, Lang J, Grandclément C, Naquin D, Ahmar M, Soulère L, et al. Lifestyle of the biotroph Agrobacterium tumefaciens in the ecological niche constructed on its host plant. New Phytol. 2018;219:350–62.
Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, et al. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci. 2018;115:E3055–E3064.
Puławska J, Kałużna M, Warabieda W, Mikiciński A. Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars-susceptible and resistant to fire blight. BMC Genom. 2017;18:868.
Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–64.
Carlier AL, Omasits U, Ahrens CH, Eberl L. Proteomics analysis of Psychotria leaf nodule symbiosis: improved genome annotation and metabolic predictions. Mol Plant Microbe Interact. 2013;26:1325–33.
Hsiao C-C, Sieber S, Georgiou A, Bailly A, Emmanouilidou D, Carlier A, et al. Synthesis and biological evaluation of the novel growth inhibitor streptol glucoside, isolated from an obligate plant symbiont. Chem A Eur J. 2018;25:1722–6.
De Mazancourt C, Loreau Mi, Dieckmann U. Understanding mutualism when there is adaptation to the partner. J Ecol. 2005;93:305–14.
Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck CC-P. Evolution of an endofungal Lifestyle: deductions from the Burkholderia rhizoxinica Genome. BMC Genomics. 2011;12:210.
Dale C, Maudlin I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol. 1999;49:267–75.
Lund MB, Kjeldsen KU, Schramm A. The earthworm-Verminephrobacter symbiosis: an emerging experimental system to study extracellular symbiosis. Front Microbiol. 2014;5:128.
Mira A, Moran NA. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 2008;44:137–43.
Greiner S, Sobanski J, Bock R. Why are most organelle genomes transmitted maternally? Bioessays. 2015;37:80–94.
Frank SA. Host–symbiont conflict over the mixing of symbiotic lineages. Proc Biol Sci. 1996;263:339–44.
Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81.
Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. Host control over infection and proliferation of a cheater symbiont. J Evol Biol. 2010;23:1919–27.
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–75.
Fraysse N, Couderc F, Poinsot V. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem. 2003;270:1365–80.
Okazaki S, Tittabutr P, Teulet A, Thouin J, Fardoux J, Chaintreuil C, et al. Rhizobium–legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS. ISME J. 2016;10:64–74.
D’Haeze W, Holsters M. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 2004;12:555–61.
Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–75.
Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–27.
Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, et al. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008;148:960–8.
Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6.
Steele MI, Kwong WK, Whiteley M, Moran NA. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. MBio. 2017;8:e01630–17.
Acknowledgements
We wish to thank Dr. Olivier Leroux (Ghent University) for his help with microscopy. This work was supported by the Flemish Fonds Wetenschappelijk Onderzoek under grant G017717N and the Special Research fund of Ghent University under grant BOF17/STA/024. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of the sequencing data. Finally, We thank three anonymous reviewers for constructive comments which substantially improved the manuscript.
Author information
Authors and Affiliations
Contributions
A.C. designed the research, F.D.M. performed the molecular biology experiments, including isolation of DNA and RNA, and phenotyping experiments. B.D. performed the genome assemblies and annotation, metagenome mining and phylogenetic analyses. T.A. provided microscopy images and microbiological data. R.R., M.T.R., and V.J. identified and collected field samples. F.D.M, B.D., and A.C. analyzed and interpreted the data; F.D.M, B.D., T.A., and A.C. wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
De Meyer, F., Danneels, B., Acar, T. et al. Adaptations and evolution of a heritable leaf nodule symbiosis between Dioscorea sansibarensis and Orrella dioscoreae. ISME J 13, 1831–1844 (2019). https://doi.org/10.1038/s41396-019-0398-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0398-8