Abstract
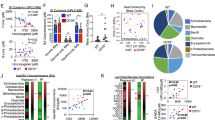
Gut microbiome community structure is associated with Crohn’s disease (CD) development and response to therapy. Bile acids (BAs) play a central role in modulating intestinal immune responses, and changes in gut bacterial communities can profoundly alter the intestinal BA pool. The liver synthesizes and conjugates primary bile acids (priBAs) that are then deconjugated, epimerized, and dehydroxylated by gut bacteria to produce secondary bile acids (secBAs). We investigated the relationship between the gut microbiome and the fecal BA pool in stool samples obtained from a well-characterized cohort of pediatric CD patients undergoing nutritional therapy to induce disease remission. We found that fecal BA composition was altered in a sub-group of CD patients who did not sustain remission. The microbial community structures associated with priBA and secBA-dominant profiles were distinct. In addition, the fecal BA concentrations were correlated with the abundance of distinct bacterial taxonomic groups. Finally, priBA dominant samples were associated with community-level decreases in enzymes for dehydroxylation but not deconjugation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The 16S rRNA gene and metagenomic sequencing data used in this study will be made available under accession PRJEB 35587 at the European Nucleotide Archive.
References
Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79.
Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–92.
Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18:489–500.
Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573.
Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301.
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6.
Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–59.
Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59.
Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–66.
Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580–5.
Humbert L, Maubert MA, Wolf C, Duboc H, Mahé M, Farabos D, et al. Bile acid profiling in human biological samples: comparison of extraction procedures and application to normal and cholestatic patients. J Chromatogr B. 2012;899:135–45.
Wells JE, Berr F, Thomas LA, Dowling RH, Hylemon PB. Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol. 2000;32:4–10.
Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7α-dehydroxylating bacteria in human feces. Clin Chim Acta. 2003;331:127–34.
Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–51.
Wahlström A, Sayin SI, Marschall H, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50.
Islam KS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–81.
Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–72.
Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PloS ONE. 2009;4:e6386.
Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–9.
Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PloS ONE. 2014;9:e115175.
Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RM, Thiele I. Personalized modeling of the human gut microbiome reveals distinct bile acid deconjugation and biotransformation potential in healthy and IBD individuals. BioRxiv. 2017:229138.
Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293.
Ruemmele F, Veres G, Kolho K, Griffiths A, Levine A, Escher J, et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J Crohn’s Colitis. 2014;8:1179–207.
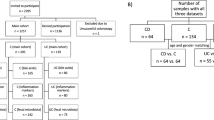
Dunn KA, Moore-Connors J, MacIntyre B, Stadnyk AW, Thomas NA, Noble A, et al. Early changes in microbial community structure are associated with sustained remission after nutritional treatment of pediatric Crohn’s disease. Inflamm Bowel Dis. 2016;22:2853–62.
MacLellan A, Connors J, Grant S, Cahill L, Langille M, Van Limbergen J. The impact of exclusive enteral nutrition (EEN) on the gut microbiome in Crohn’s disease: a review. Nutrients. 2017;9:447.
Turner D, Griffiths AM, Walters TD, Seah T, Markowitz J, Pfefferkorn M, et al. Mathematical weighting of the pediatric Crohn’s disease activity index (PCDAI) and comparison with its other short versions. Inflamm Bowel Dis. 2011;18:55–62.
Shafiei M, Dunn KA, Boon E, MacDonald SM, Walsh DA, Gu H, et al. BioMiCo: a supervised Bayesian model for inference of microbial community structure. Microbiome. 2015;3:8.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’hara R, et al. vegan: Community Ecology Package.R package version 2.3–0.2015. 2017.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B. 1995;57:289–300.
Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902.
Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534–41.
Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–7.
Narushima S, Itoh K, Miyamoto Y, Park S, Nagata K, Kuruma K, et al. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41:835–43.
Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–55.
Brown JR, Flemer B, Joyce SA, Zulquernain A, Sheehan D, Shanahan F, et al. Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for Clostridioides difficile infection. BMC Gastroenterol. 2018;18:131.
Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. 2015;11:685.
Harris SC, Devendran S, Méndez-García C, Mythen SM, Wright CL, Fields CJ, et al. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243T. Gut Microbes. 2018;9:523–39.
Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, et al. Accessible, curated metagenomic data through ExperimentHub. Nat Methods. 2017;14:1023.
Martin G, Kolida S, Marchesi J, Want E, Sidaway J, Swann JR. In vitro modeling of bile acid processing by the human fecal microbiota. Front Microbiol. 2018;9:1153.
Martínez-Moya P, Romero-Calvo I, Requena P, Hernández-Chirlaque C, Aranda CJ, González R, et al. Dose-dependent antiinflammatory effect of ursodeoxycholic acid in experimental colitis. Int Immunopharmacol. 2013;15:372–80.
Ward JB, Lajczak NK, Kelly OB, O’Dwyer AM, Giddam AK, Ní Gabhann J, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol-Gastrointest Liver Physiol. 2017;312:G550–8.
Hirano S, Masuda N. Epimerization of the 7-hydroxy group of bile acids by the combination of two kinds of microorganisms with 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activity, respectively. J Lipid Res. 1981;22:1060–8.
Akao T, Akao T, Kobashi K. Purification and characterization of 7β-Hydroxysteroid dehydrogenase from Ruminococus sp. of human intestine. J Biochem. 1987;102:613–9.
Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7beta-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res. 2013;54:3062–9.
Lepercq P, Gérard P, Béguet F, Grill J, Relano P, Cayuela C, et al. Isolates from normal human intestinal flora but not lactic acid bacteria exhibit 7α-and 7β-hydroxysteroid dehydrogenase activities. Micro Ecol Health Dis. 2004;16:195–201.
Pols TW, Puchner T, Korkmaz HI, Vos M, Soeters MR, de Vries CJ. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PloS ONE. 2017;12:e0176715.
Liu J, Yan Q, Luo F, Shang D, Wu D, Zhang H, et al. Acute cholecystitis associated with infection of Enterobacteriaceae from gut microbiota. Clin Microbiol Infect. 2015;21:851. e1–9.
Buttó LF, Schaubeck M, Haller D. Mechanisms of microbe–host interaction in Crohn’s disease: dysbiosis vs. pathobiont selection. Front Immunol. 2015;6:555.
Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103.
Acknowledgements
The authors would like to thank all participating children and their families, as well as the pediatric dietitians (Jennifer Haskett, Lisa Parkinson-McGraw) and the IWK GI-Research Team (Brad MacIntyre). JVL was supported by a Canadian Institutes of Health Research (CIHR)-Canadian Association of Gastroenterology-Crohn’s Colitis Canada New Investigator Award (2015–2019), a Canada Research Chair Tier 2 in Translational Microbiomics (2018–2019) and a Canadian Foundation of Innovation John R. Evans Leadership fund (awards #35235 and #36764), a Nova Scotia Health Research Foundation (NSHRF) establishment award (2015–2017), an IWK Health Centre Research Associateship (for JC), an IWK Health Centre Project grant (Cat. B—2017), a American Gastroenterology Association Pfizer Young Investigator Pilot Research Award in Inflammatory Bowel Disease (2018), a donation from the MacLeod family and by a CIHR-SPOR-Chronic Diseases grant (Inflammation, Microbiome, and Alimentation: Gastro-Intestinal and Neuropsychiatric Effects: the IMAGINE-SPOR chronic disease network). JVL reports consulting, travel and/or speaker fees and research support from AbbVie, Janssen, Nestlé Health Science, Merck, P&G, GSK, Illumina, Otsuka.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
JVL reports consulting, travel and/or speaker fees and research support from AbbVie, Janssen, Nestlé Health Science, Merck, P&G, GSK, Illumina, Otsuka. The other authors have no conflicts to report.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised: Jessica Connors, Katherine Dunn
Rights and permissions
About this article
Cite this article
Connors, J., Dunn, K.A., Allott, J. et al. The relationship between fecal bile acids and microbiome community structure in pediatric Crohn’s disease. ISME J 14, 702–713 (2020). https://doi.org/10.1038/s41396-019-0560-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0560-3
This article is cited by
-
Comparative metagenomics reveals the differential gut microbiota involved in bile acid metabolism in patients with crohn’s disease
World Journal of Microbiology and Biotechnology (2026)
-
Disruption of gut microbiome and metabolome in treatment-naïve children with attention deficit hyperactivity disorder
BMC Microbiology (2025)
-
Targeted LC-MS/MS profiling of bile acids reveals primary/secondary bile acid ratio as a novel biomarker for necrotizing enterocolitis
Analytical and Bioanalytical Chemistry (2024)
-
Defecation status, intestinal microbiota, and habitual diet are associated with the fecal bile acid composition: a cross-sectional study in community-dwelling young participants
European Journal of Nutrition (2023)
-
Clostridioides difficile Infection in Pediatric Inflammatory Bowel Disease
Current Gastroenterology Reports (2023)