Abstract
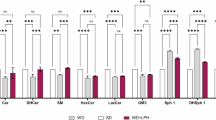
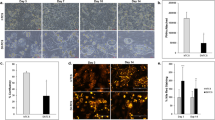
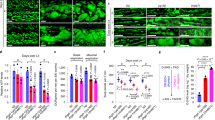
3,3′,4′,5,7-Pentahydroxyflavone-3-rhamnoglucoside (rutin) is a flavonoid with a wide range of pharmacological activities. Dietary rutin is hardly absorbed because the microflora in the large intestine metabolize rutin into a variety of compounds including quercetin and phenol derivatives such as 3,4-dihydroxyphenolacetic acid (DHPAA), 3,4-dihydroxytoluene (DHT), 3,4-hydroxyphenylacetic acid (HPAA) and homovanillic acid (HVA). We examined the potential of rutin and its metabolites as novel histone acetyltransferase (HAT) inhibitors. DHPAA, HPAA and DHT at the concentration of 25 μM significantly inhibited in vitro HAT activity with DHT having the strongest inhibitory activity. Furthermore, DHT was shown to be a highly efficient inhibitor of p300 HAT activity, which corresponded with its high degree of inhibition on intracellular lipid accumulation in HepG2 cells. Docking simulation revealed that DHT was bound to the p300 catalytic pocket, bromodomain. Drug affinity responsive target stability (DARTS) analysis further supported the possibility of direct binding between DHT and p300. In HepG2 cells, DHT concentration-dependently abrogated p300-histone binding and induced hypoacetylation of histone subunits H3K9, H3K36, H4K8 and H4K16, eventually leading to the downregulation of lipogenesis-related genes and attenuating lipid accumulation. In ob/ob mice, administration of DHT (10, 20 mg/kg, iv, every other day for 6 weeks) dose-dependently improved the NAFLD pathogenic features including body weight, liver mass, fat mass, lipid accumulation in the liver, and biochemical blood parameters, accompanied by the decreased mRNA expression of lipogenic genes in the liver. Our results demonstrate that DHT, a novel p300 histone acetyltransferase inhibitor, may be a potential preventive or therapeutic agent for NAFLD.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Shankar E, Kanwal R, Candamo M, Gupta S. Dietary phytochemicals as epigenetic modifiers in cancer: promise and challenges. Semin Cancer Biol. 2016;40–41:82–99.
Fotbolcu H, Zorlu E. Nonalcoholic fatty liver disease as a multi-systemic disease. World J Gastroenterol. 2016;22:4079–90.
Sun C, Fan JG, Qiao L. Potential epigenetic mechanism in non-alcoholic fatty liver disease. Int J Mol Sci. 2015;16:5161–79.
Tariq A, Mussarat S, Adnan M. Review on ethnomedicinal, phytochemical and pharmacological evidence of Himalayan anticancer plants. J Ethnopharmacol. 2015;164:96–119.
Choi WJ, Kim SK, Park HK, Sohn UD, Kim W. Anti-inflammatory and anti-superbacterial properties of sulforaphane from shepherd’s purse. Korean J Physiol Pharmacol. 2014;18:33–9.
Lee Y-H, Kwak J, Choi H-K, Choi K-C, Kim S, Lee J, et al. EGCG suppresses prostate cancer cell growth modulating acetylation of androgen receptor by anti-histone acetyltransferase activity. Int J Mol Med. 2012;30:69–74.
Shankar S, Kumar D, Srivastava RK. Epigenetic modifications by dietary phytochemicals: implications for personalized nutrition. Pharmacol Ther. 2013;138:1–17.
Wang G-L, Salisbury E, Shi X, Timchenko L, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPα in the inhibition of liver proliferation in old mice. J Biol Chem. 2008;283:26169–78.
Bose M, Lambert JD, Ju J, Reuhl KR, Shapses SA, Yang CS. The major green tea polyphenol,(-)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat–fed mice. J Nutr. 2008;138:1677–83.
Chen YK, Cheung C, Reuhl KR, Liu AB, Lee MJ, Lu YP, et al. Effects of green tea polyphenol (−)-epigallocatechin-3-gallate on newly developed high-fat/western-style diet-induced obesity and metabolic syndrome in mice. J Agric Food Chem. 2011;59:11862–71.
Chung MY, Song JH, Lee J, Shin EJ, Park JH, Lee SH, et al. Tannic acid, a novel histone acetyltransferase inhibitor, prevents non-alcoholic fatty liver disease both in vivo and in vitro model. Mol Metab. 2019;19:34–48.
Asrih M, Jornayvaz FR. Inflammation as a potential link between nonalcoholic fatty liver disease and insulin resistance. J Endocrinol. 2013;218:R25–36.
Kitade H, Chen G, Ni Y, Ota T. Nonalcoholic fatty liver disease and insulin resistance: new insights and potential new treatments. Nutrients. 2017;9:387.
Li Z, Li Y, Zhang HX, Guo JR, Lam CWK, Wang CY, et al. Mitochondria‐mediated pathogenesis and therapeutics for non‐alcoholic fatty liver disease. Mol Nutr Food Res. 2019;63:1900043.
Masarone M, Rosato V, Dallio M, Gravina AG, Aglitti A, Loguercio C, et al. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. Oxid Med Cell Longev. 2018;2018:7864316.
Simões IC, Fontes A, Pinton P, Zischka H, Wieckowski MR. Mitochondria in non-alcoholic fatty liver disease. Int J Biochem Cell Biol. 2018;95:93–9.
Al-Dhabi NA, Arasu MV, Park CH, Park SU. An up-to-date review of rutin and its biological and pharmacological activities. EXCLI J. 2015;14:59–63.
Sharma S, Ali A, Ali J, Sahni JK, Baboota S. Rutin: therapeutic potential and recent advances in drug delivery. Expert Opin Investig Drugs. 2013;22:1063–79.
Jaganath IB, Jaganath IB, Mullen W, Edwards CA, Crozier A. The relative contribution of the small and large intestine to the absorption and metabolism of rutin in man. Free Radic Res. 2006;40:1035–46.
Su KY, Yu CY, Chen YP, Hua KF, Chen YLS. 3, 4-Dihydroxytoluene, a metabolite of rutin, inhibits inflammatory responses in lipopolysaccharide-activated macrophages by reducing the activation of NF-κB signaling. BMC Complement Alter Med. 2014;14:21.
Morales AM, Mukai R, Murota K, Terao J. Inhibitory effect of catecholic colonic metabolites of rutin on fatty acid hydroperoxide and hemoglobin dependent lipid peroxidation in Caco-2 cells. J Clin Biochem Nutr. 2018;63:175–80.
Giménez‐Bastida JA, Zielinski H, Piskula M, Zielinska D, Szawara‐Nowak D. Buckwheat bioactive compounds, their derived phenolic metabolites and their health benefits. Mol Nutr Food Res. 2017;61:1600475.
Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, et al. Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci U S A. 2009;106:21984–9.
Sharma R, Zhou MM. Partners in crime: The role of tandem modules in gene transcription. Protein Sci. 2015;24:1347–59.
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–33.
Manning ET, Ikehara T, Ito T, Kadonaga JT, Kraus WL. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol Cell Biol. 2001;21:3876–87.
da Silva RP, Kelly KB, Al Rajabi A, Jacobs RL. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40:277–83.
Lu SC, Alvarez L, Huang Z-Z, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98:5560–5.
Sinton MC, Hay DC, Drake AJ. Metabolic control of gene transcription in non-alcoholic fatty liver disease: the role of the epigenome. Clin Epigenet. 2019;11:104.
Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–9.
Ferreira DM, Simão AL, Rodrigues CM, Castro RE. Revisiting the metabolic syndrome and paving the way for micro RNA s in non‐alcoholic fatty liver disease. FEBS J. 2014;281:2503–24.
Legeay S, Rodier M, Fillon L, Faure S, Clere N. Epigallocatechin gallate: a review of its beneficial properties to prevent metabolic syndrome. Nutrients. 2015;7:5443–68.
Shanak S, Saad B, Zaid H. Metabolic and epigenetic action mechanisms of antidiabetic medicinal plants. Evid Based Complement Alternat Med. 2019;2019:3583067.
Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278:19134–40.
Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–74.
Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4.
Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102.
De Long NE, Hardy DB, Ma N, Holloway AC. Increased incidence of non‐alcoholic fatty liver disease in male rat offspring exposed to fluoxetine during fetal and neonatal life involves the NLRP3 inflammasome and augmented de novo hepatic lipogenesis. J Appl Toxicol. 2017;37:1507–16.
Chung S, Hwang JT, Park JH, Choi HK. Free fatty acid-induced histone acetyltransferase activity accelerates lipid accumulation in HepG2 cells. Nutr Res Pr. 2019;13:196–204.
Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. p300 interacts with the N-and C-terminal part of PPARγ2 in a ligand-independent and-dependent manner, respectively. J Biol Chem. 1999;274:7681–8.
Giandomenico V, Simonsson M, Grönroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23:2587–99.
Tian Y, Wong VWS, Chan HLY, Cheng ASL. Epigenetic regulation of hepatocellular carcinoma in non-alcoholic fatty liver disease. Semin Cancer Biol 2013;23:471–82.
Maksimoska J, Segura-Peña D, Cole PA, Marmorstein R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry. 2014;53:3415–22.
Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–6.
Cochran AG, Conery AR, Sims RJ. Bromodomains: a new target class for drug development. Nat Rev Drug Discov. 2019;18:609–28.
Chang L, Takada S. Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations. Sci Rep. 2016;6:34441.
Zhang F, Huang Q, Yan J, Chen Z. Histone acetylation induced transformation of B-DNA to Z-DNA in cells probed through FT-IR spectroscopy. Anal Chem. 2016;88:4179–82.
Acknowledgements
This study was supported by the Main Research Program (E-0150301) of the Korea Food Research Institute (KFRI), funded by the Ministry of Science, ICT & Future Planning.
Author information
Authors and Affiliations
Contributions
HKC and JTH designed the research. JL, JHS, MYC, and HKC performed the research. JL, JHL, TGN, and JHP contributed to the data analysis. JL, HKC, and JTH wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, J., Song, JH., Chung, MY. et al. 3,4-dihydroxytoluene, a metabolite of rutin, suppresses the progression of nonalcoholic fatty liver disease in mice by inhibiting p300 histone acetyltransferase activity. Acta Pharmacol Sin 42, 1449–1460 (2021). https://doi.org/10.1038/s41401-020-00571-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-020-00571-7
Keywords
This article is cited by
-
The influence of Mg(II) and Ca(II) ions on the autoxidation of 4-methylcatechol in weakly alkaline aqueous solutions
Reaction Kinetics, Mechanisms and Catalysis (2022)