Abstract
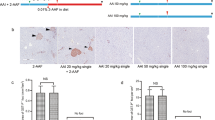
Aristolochic acids (AAs) have been identified as a significant risk factor for hepatocellular carcinoma (HCC). Ferroptosis is a type of regulated cell death involved in the tumor development. In this study, we investigated the molecular mechanisms by which AAs enhanced the growth of HCC. By conducting bioinformatics and RNA-Seq analyses, we found that AAs were closely correlated with ferroptosis. The physical interaction between p53 and AAs in HepG2 cells was validated by bioinformatics analysis and SPR assays with the binding pocket sites containing Pro92, Arg174, Asp207, Phe212, and His214 of p53. Based on the binding pocket that interacts with AAs, we designed a mutant and performed RNA-Seq profiling. Interestingly, we found that the binding pocket was responsible for ferroptosis, GADD45A, NRF2, and SLC7A11. Functionally, the interaction disturbed the binding of p53 to the promoter of GADD45A or NRF2, attenuating the role of p53 in enhancing GADD45A and suppressing NRF2; the mutant did not exhibit the same effects. Consequently, this event down-regulated GADD45A and up-regulated NRF2, ultimately inhibiting ferroptosis, suggesting that AAs hijacked p53 to down-regulate GADD45A and up-regulate NRF2 in HepG2 cells. Thus, AAs treatment resulted in the inhibition of ferroptosis via the p53/GADD45A/NRF2/SLC7A11 axis, which led to the enhancement of tumor growth. In conclusion, AAs-hijacked p53 restrains ferroptosis through the GADD45A/NRF2/SLC7A11 axis to enhance tumor growth. Our findings provide an underlying mechanism by which AAs enhance HCC and new insights into p53 in liver cancer. Therapeutically, the oncogene NRF2 is a promising target for liver cancer.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The RNA-Seq data are available at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/), accession number: PRJNA1000259.
References
Lu ZN, Luo Q, Zhao LN, Shi Y, Wang N, Wang L, et al. The mutational features of aristolochic acid-induced mouse and human liver cancers. Hepatology. 2020;71:929–42.
Nault JC, Letouzé E. Mutational processes in hepatocellular carcinoma: the story of aristolochic acid. Semin Liver Dis. 2019;39:334–40.
Chen S, Dong Y, Qi X, Cao Q, Luo T, Bai Z, et al. Aristolochic acids exposure was not the main cause of liver tumorigenesis in adulthood. Acta Pharm Sin B. 2022;12:2252–67.
Das S, Thakur S, Korenjak M, Sidorenko VS, Chung FF, Zavadil J. Aristolochic acid-associated cancers: a public health risk in need of global action. Nat Rev Cancer. 2022;22:576–91.
Hu J, Cao J, Topatana W, Juengpanich S, Li S, Zhang B, et al. Targeting mutant p53 for cancer therapy: direct and indirect strategies. J Hematol Oncol. 2021;14:157–76.
Stiewe T, Haran TE. How mutations shape p53 interactions with the genome to promote tumorigenesis and drug resistance. Drug Resist Updat. 2018;38:27–43.
Morris JPT, Yashinskie JJ, Koche R, Chandwani R, Tian S, Chen CC, et al. α-Ketoglutarate links p53 to cell fate during tumour suppression. Nature. 2019;573:595–9.
Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71:616–30.
Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363–74.
Makino Y, Hikita H, Fukumoto K, Sung JH, Sakano Y, Murai K, et al. Constitutive activation of the tumor suppressor p53 in hepatocytes paradoxically promotes non-cell autonomous liver carcinogenesis. Cancer Res. 2022;82:2860–73.
Kim J, Yu L, Chen W, Xu Y, Wu M, Todorova D, et al. Wild-type p53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative phosphorylation. Cancer Cell. 2019;35:191–203.
Pietrasik S, Zajac G, Morawiec J, Soszynski M, Fila M, Blasiak J. Interplay between BRCA1 and GADD45A and its potential for nucleotide excision repair in breast cancer pathogenesis. Int J Mol Sci. 2020;21:870–92.
Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–80.
Dodson M, Castro-Portuguez R, Zhang DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:101–7.
Kudo Y, Sugimoto M, Arias E, Kasashima H, Cordes T, Linares JF, et al. PKCλ/ι loss induces autophagy, oxidative phosphorylation, and NRF2 to promote liver cancer progression. Cancer Cell. 2020;38:247–62.
Lu C, Xue L, Luo K, Liu Y, Lai J, Yao X, et al. Colon-accumulated gold nanoclusters alleviate intestinal inflammation and prevent secondary colorectal carcinogenesis via nrf2-dependent macrophage reprogramming. ACS Nano. 2023;17:18421–32.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–97.
Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18:280–96.
Lyu N, Zeng Y, Kong Y, Chen Q, Deng H, Ou S, et al. Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann Transl Med. 2021;9:675–86.
Wang YF, Feng JY, Zhao LN, Zhao M, Wei XF, Geng Y, et al. Aspirin triggers ferroptosis in hepatocellular carcinoma cells through restricting NF-κB p65-activated SLC7A11 transcription. Acta Pharmacol Sin. 2023;44:1712–24.
Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620.
Louandre C, Marcq I, Bouhlal H, Lachaier E, Godin C, Saidak Z, et al. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015;356:971–7.
Wang X, Chen X, Zhou W, Men H, Bao T, Sun Y, et al. Ferroptosis is essential for diabetic cardiomyopathy and is prevented by sulforaphane via AMPK/NRF2 pathways. Acta Pharm Sin B. 2022;12:708–22.
Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054–81.
Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62.
Ma WQ, Sun XJ, Zhu Y, Liu NF. Metformin attenuates hyperlipidaemia-associated vascular calcification through anti-ferroptotic effects. Free Radic Biol Med. 2021;165:229–42.
Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580:93–9.
Wu J, Feng Z, Chen L, Li Y, Bian H, Geng J, et al. TNF antagonist sensitizes synovial fibroblasts to ferroptotic cell death in collagen-induced arthritis mouse models. Nat Commun. 2022;13:676–92.
Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, et al. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551:639–43.
Yang Y, Luo M, Zhang K, Zhang J, Gao T, Connell DO, et al. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat Commun. 2020;11:433–47.
Liu Z, Wu J, Cai C, Yang B, Qi ZM. Flexible hyperspectral surface plasmon resonance microscopy. Nat Commun. 2022;13:6475–90.
Li G, Kryczek I, Nam J, Li X, Li S, Li J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23:526–37.
Poon SL, Huang MN, Choo Y, McPherson JR, Yu W, Heng HL, et al. Mutation signatures implicate aristolochic acid in bladder cancer development. Genome Med. 2015;7:38–48.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–w64.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–d12.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. 2020;2020:baaa021–8.
Crampon K, Giorkallos A, Deldossi M, Baud S, Steffenel LA. Machine-learning methods for ligand-protein molecular docking. Drug Discov Today. 2022;27:151–64.
Chang YS, Tu SJ, Chen HD, Hsu MH, Chen YC, Chao DS, et al. Integrated genomic analyses of hepatocellular carcinoma. Hepatol Int. 2023;17:97–111.
Lertvachirapaiboon C, Baba A, Ekgasit S, Shinbo K, Kato K, Kaneko F. Transmission surface plasmon resonance techniques and their potential biosensor applications. Biosens Bioelectron. 2018;99:399–415.
Wei RJ, Wu WR, Pan CT, Yu CY, Li CF, Chen LR, et al. Inhibition of the formation of autophagosome but not autolysosome augments ABT-751-induced apoptosis in TP53-deficient Hep-3B cells. J Cell Physiol. 2019;234:9551–63.
Tian R, Abarientos A, Hong J, Hashemi SH, Yan R, Dräger N, et al. Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat Neurosci. 2021;24:1020–34.
Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov. 2023;22:127–44.
Fiore A, Zeitler L, Russier M, Groß A, Hiller MK, Parker JL, et al. Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling. Mol Cell. 2022;82:920–32.
Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, et al. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–84.
Wang Y, Wang Z, Wu Z, Chen M, Dong D, Yu P, et al. Involvement of REV-ERBα dysregulation and ferroptosis in aristolochic acid I-induced renal injury. Biochem Pharmacol. 2021;193:114807–12.
Gautheron J, Luedde T. A novel player in inflammation and cancer: the deubiquitinase CYLD controls HCC development. J Hepatol. 2012;57:937–9.
Li R, Shugart YY, Zhou W, An Y, Yang Y, Zhou Y, et al. Common genetic variations of the cytochrome P450 1A1 gene and risk of hepatocellular carcinoma in a Chinese population. Eur J Cancer. 2009;45:1239–47.
Gulve N, Su C, Deng Z, Soldan SS, Vladimirova O, Wickramasinghe J, et al. DAXX-ATRX regulation of p53 chromatin binding and DNA damage response. Nat Commun. 2022;13:5033–47.
Charette N, De Saeger C, Lannoy V, Horsmans Y, Leclercq I, Stärkel P. Salirasib inhibits the growth of hepatocarcinoma cell lines in vitro and tumor growth in vivo through ras and mTOR inhibition. Mol Cancer. 2010;9:256–14.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82372818 to Xiao-dong Zhang; 82103066 to Guang Yang; 82302887 to Hong-feng Yuan; 82303210 to Yu-fei Wang).
Author information
Authors and Affiliations
Contributions
XDZ, GY, and CYH concepted and designed the project; YHS and HFY Developed the methodology; CYH acquired the data Acquisition of data (provided animals, provided facilities, etc.); CYH, LNZ, YFW, HHZ, LLS, JS, WL and PL analyzed and interpreted the data (e.g., statistical analysis, biostatistics, computational analysis); CYH, NNZ and XDZ written, reviewed, and/or revised of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hou, Cy., Suo, Yh., Lv, P. et al. Aristolochic acids-hijacked p53 promotes liver cancer cell growth by inhibiting ferroptosis. Acta Pharmacol Sin 46, 208–221 (2025). https://doi.org/10.1038/s41401-024-01354-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-024-01354-0
Keywords
This article is cited by
-
Bone marrow–derived mesenchymal stem cells alleviate hepatic lipid metabolism disorders after scald injury: integrating liver transcriptome and metabolome
Stem Cell Research & Therapy (2026)
-
Lactylation-mediated miRNA regulation in cancer therapy resistance
Journal of Translational Medicine (2025)
-
Redox mechanism of glycerophospholipids and relevant targeted therapy in ferroptosis
Cell Death Discovery (2025)
-
LZTS2 methylation as a potential diagnostic and prognostic marker in LIHC and STAD: Evidence from bioinformatics and in vitro analyses
Scientific Reports (2025)
-
CD155 promotes the advancement of hepatocellular carcinoma by suppressing the p53-mediated ferroptosis via interacting with CD96
Journal of Molecular Medicine (2025)