Abstract
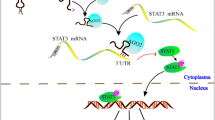
Gastric cancer is a malignant gastrointestinal disease characterized by high morbidity and mortality rates worldwide. The occurrence and progression of gastric cancer are influenced by various factors, including the abnormal alternative splicing of key genes. Recently, RBM39 has emerged as a tumor biomarker that regulates alternative splicing in several types of cancer. However, the specific functions and key alternative splicing events modulated by RBM39 in gastric cancer are still unclear. In this work, bioinformatic analysis of The Cancer Genome Atlas (TCGA) database and immunoblotting of patient tissue samples revealed that RBM39 was highly expressed in gastric cancer tissues and that its elevated expression significantly reduced overall patient survival. Cell-line-based and tumor xenograft experiments demonstrated that RBM39 knockdown attenuated the growth of gastric cancer cells both in vitro and in vivo. Mechanistically, through RNA-seq, minigene, and RT‒PCR, we discovered and further validated that RBM39 inhibited exon 3 skipping, thereby modulating the splicing of MRPL33. The long isoform MRPL33-L, which includes exon 3, but not the short isoform MRPL33-S, which lacks exon 3, significantly promoted the proliferation and colony formation of gastric cancer cells. Furthermore, we observed an increased percent-splice-in (PSI) of MRPL33 in gastric cancer tissues. Genetic manipulation and pharmacological treatment with the RBM39 degrader indisulam demonstrated that RBM39 regulated cell proliferation by influencing the splicing switch of MRPL33 in gastric cancer cells and a xenograft mouse model. Our findings indicate that RBM39 regulates the oncogenic splicing of MRPL33 and suggest that it may serve as a potential therapeutic target for gastric cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48.
Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–41.
Li W, Deng X, Chen J. RNA-binding proteins in regulating mRNA stability and translation: roles and mechanisms in cancer. Semin Cancer Biol. 2022;86:664–77.
Xu Y, Nijhuis A, Keun HC. RNA-binding motif protein 39 (RBM39): an emerging cancer target. Br J Pharmacol. 2022;179:2795–812.
Huang SC, Zhang HS, Yu B, McMahon E, Nguyen DT, Yu FH, et al. Protein 4.1R exon 16 3’ splice site activation requires coordination among TIA1, Pcbp1, and RBM39 during terminal erythropoiesis. Mol Cell Biol. 2017;37:e00446–16.
Eléouët M, Lu C, Zhou Y, Yang P, Ma J, Xu G. Insights on the biological functions and diverse regulation of RNA-binding protein 39 and their implication in human diseases. Biochim Biophys Acta Gene Regul Mech. 2023;1866:194902.
Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, et al. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol Cell. 2005;17:429–39.
Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem. 2002;277:1229–34.
Dutta J, Fan G, Gelinas C. CAPERα is a novel Rel-TAD-interacting factor that inhibits lymphocyte transformation by the potent Rel/NF-κB oncoprotein v-Rel. J Virol. 2008;82:10792–802.
Kang YK, Putluri N, Maity S, Tsimelzon A, Ilkayeva O, Mo Q, et al. CAPER is vital for energy and redox homeostasis by integrating glucose-induced mitochondrial functions via ERR-α-Gabpa and stress-induced adaptive responses via NF-κB-cMYC. PLoS Genet. 2015;11:e1005116.
Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell. 2019;35:369–84.
Hsiehchen D, Goralski M, Kim J, Xie Y, Nijhawan D. Biomarkers for RBM39 degradation in acute myeloid leukemia. Leukemia. 2020;34:1924–8.
Zhang X, Yang L, Liu X, Nie Z, Liu M, Wang T, et al. Regulatory role of RBM39 in acute myeloid leukemia: mediation through the PI3K/AKT pathway. Biochim Biophys Acta Mol Cell Res. 2024;1871:119607.
Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:103025.
Ting TC, Goralski M, Klein K, Wang B, Kim J, Xie Y, et al. Aryl sulfonamides degrade RBM39 and RBM23 by recruitment to CRL4-DCAF15. Cell Rep. 2019;29:1499–510.
Chai Y, Liu X, Dai L, Li Y, Liu M, Zhang JY. Overexpression of HCC1/CAPERα may play a role in lung cancer carcinogenesis. Tumour Biol. 2014;35:6311–7.
Mercier I, Gonzales DM, Quann K, Pestell TG, Molchansky A, Sotgia F, et al. CAPER, a novel regulator of human breast cancer progression. Cell Cycle. 2014;13:1256–64.
Faust TB, Yoon H, Nowak RP, Donovan KA, Li Z, Cai Q, et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat Chem Biol. 2020;16:7–14.
Lu J, Jiang H, Li D, Chen T, Wang Y, Pu Z, et al. Proximity labeling, quantitative proteomics, and biochemical studies revealed the molecular mechanism for the inhibitory effect of indisulam on the proliferation of gastric cancer cells. J Proteome Res. 2021;20:4462–74.
Liu L, Luo C, Luo Y, Chen L, Liu Y, Wang Y, et al. MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene. 2018;37:86–94.
Li J, Feng D, Gao C, Zhang Y, Xu J, Wu M, et al. Isoforms S and L of MRPL33 from alternative splicing have isoform‑specific roles in the chemoresponse to epirubicin in gastric cancer cells via the PI3K/AKT signaling pathway. Int J Oncol. 2019;54:1591–600.
Hou C, Wu X, Shi R, Xing X, Tian S, Eléouët M, et al. Subtle structural alteration in indisulam switches the molecular mechanisms for the inhibitory effect on the migration of gastric cancer cells. Biomed Pharmacother. 2024;172:116259.
Zhang S, Wang Y, Cao Y, Wu J, Zhang Z, Ren H, et al. Inhibition of the PINK1-Parkin pathway enhances the lethality of sorafenib and regorafenib in hepatocellular carcinoma. Front Pharmacol. 2022;13:851832.
Yu W, Wang B, Zhou L, Xu G. Endoplasmic reticulum stress-mediated p62 downregulation inhibits apoptosis via c-Jun upregulation. Biomol Ther (Seoul). 2021;29:195–204.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:884–90.
Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–30.
Xu C, Chen X, Zhang X, Zhao D, Dou Z, Xie X, et al. RNA-binding protein 39: A promising therapeutic target for cancer. Cell Death Discov. 2021;7:214.
Chen WC, To MD, Westcott PMK, Delrosario R, Kim IJ, Philips M, et al. Targeting KRAS4A splicing through the RBM39/DCAF15 pathway inhibits cancer stem cells. Nat Commun. 2021;12:4288.
Mai S, Qu X, Li P, Ma Q, Cao C, Liu X. Global regulation of alternative RNA splicing by the SR-rich protein RBM39. Biochim Biophys Acta. 2016;1859:1014–24.
Singh S, Quarni W, Goralski M, Wan S, Jin H, Van de Velde LA, et al. Targeting the spliceosome through RBM39 degradation results in exceptional responses in high-risk neuroblastoma models. Sci Adv. 2021;7:eabj5405.
Wang J, Su W, Zhang T, Zhang S, Lei H, Ma F, et al. Aberrant Cyclin D1 splicing in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2023;14:244.
Zhang R, Wang W, Zhang N, Chen X, Liu W, Zhang L, et al. Systematic pan-cancer analysis identifies RBM39 as an immunological and prognostic biomarker. J Cell Mol Med. 2022;26:4859–71.
Nijhuis A, Sikka A, Yogev O, Herendi L, Balcells C, Ma Y, et al. Indisulam targets RNA splicing and metabolism to serve as a therapeutic strategy for high-risk neuroblastoma. Nat Commun. 2022;13:1380.
Puvvula PK, Yu Y, Sullivan KR, Eyob H, Rosenberg J, Welm A, et al. Inhibiting an RBM39/MLL1 epigenomic regulatory complex with dominant-negative peptides disrupts cancer cell transcription and proliferation. Cell Rep. 2021;35:109156.
Xu Y, Spear S, Ma Y, Lorentzen MP, Gruet M, McKinney F, et al. Pharmacological depletion of RNA splicing factor RBM39 by indisulam synergizes with PARP inhibitors in high-grade serous ovarian carcinoma. Cell Rep. 2023;42:113307.
Campagne S, Jutzi D, Malard F, Matoga M, Romane K, Feldmuller M, et al. Molecular basis of RNA-binding and autoregulation by the cancer-associated splicing factor RBM39. Nat Commun. 2023;14:5366.
Bhattacharyya A, Trotta CR, Narasimhan J, Wiedinger KJ, Li W, Effenberger KA, et al. Small molecule splicing modifiers with systemic HTT-lowering activity. Nat Commun. 2021;12:7299.
Allegri L, Baldan F, Roy S, Aube J, Russo D, Filetti S, et al. The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells. Sci Rep. 2019;9:7374.
Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, et al. HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells. Sci Rep. 2017;7:9694.
Deymeer F. Nusinersen in SMA 2 and 3: risks vs benefits. Neurology. 2020;95:151–2.
Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–48.
Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–44.
Acknowledgements
We would like to express our gratitude to Prof. Ying Feng at the Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, for generously providing the MRPL33 minigene plasmids. This work was supported by the National Natural Science Foundation of China (32171437), the National Key R&D Program of China (2019YFA0802401), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (22KJB310017), Suzhou Science and Technology Development Project (SZM2023006), Gusu Key Health Talent Program of Suzhou (GSWS2022122), the Young Physician Scientists Program (ML42900323) and Interdisciplinary Basic Frontier Innovation Program (YXY2302015) at Suzhou Medical College of Soochow University, the National Center for International Research (2017B01012), the Jiangsu Key Laboratory of Neuropsychiatric Diseases (BM2013003), and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Contributions
CPL, JBL, XGJ, JJM, and GX designed the research; CPL performed the research, DBL and YHW collected patient samples; CPL and GX analyzed the data and drafted the paper; all authors revised the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, Cp., Li, Jb., Li, Db. et al. RNA-binding motif protein RBM39 enhances the proliferation of gastric cancer cells by facilitating an oncogenic splicing switch in MRPL33. Acta Pharmacol Sin 46, 1068–1081 (2025). https://doi.org/10.1038/s41401-024-01431-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-024-01431-4