Abstract
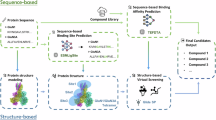
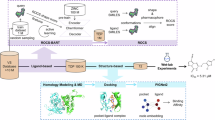
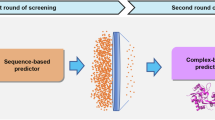
N-methyl-D-aspartate receptors (NMDARs) are critical mediators of excitatory neurotransmission and are composed of seven subunits (GluN1, GluN2A–D, and GluN3A–B) that form diverse receptor subtypes. While GluN1/GluN2 subtypes have been extensively characterized and have led to approved therapeutics, the GluN1/GluN3A subtype remains underexplored despite emerging evidence of its involvement in neuropsychiatric disorders. Efficient identification of modulators requires accurate prediction of drug-target affinity (DTA), particularly for challenging targets such as GluN1/GluN3A. In this study, we applied the ImageDTA model, which is a multiscale 2D convolutional neural network (CNN), to virtually screen 18 million small molecules for GluN1/GluN3A inhibitors. This artificial intelligence (AI)-driven approach prioritized 12 compounds, three of which demonstrated potent inhibitory activity (IC₅₀ < 30 µM) in experimental validation. The most potent hit, with an IC50 of 4.16 ± 0.65 µM, revealed a novel structural scaffold, thus highlighting the potential of AI in accelerating drug discovery for underexplored receptor subtypes. These findings establish a robust framework for advancing GluN1/GluN3A-targeted therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol Rev. 2021;73:1469–658.
Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400.
Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol. 2015;20:14–23.
Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, et al. Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol. 2018;150:1081–105.
Egunlusi AO, Joubert J. NMDA receptor antagonists: emerging insights into molecular mechanisms and clinical applications in neurological disorders. Pharmaceuticals. 2024;17:639.
Ahmed H, Haider A, Ametamey SM. N-Methyl-D-Aspartate (NMDA) receptor modulators: a patent review (2015-present). Expert Opin Ther Pat. 2020;30:743–67.
Bossi S, Pizzamiglio L, Paoletti P. Excitatory GluN1/GluN3A glycine receptors (eGlyRs) in brain signaling. Trends Neurosci. 2023;46:667–81.
Bossi S, Dhanasobhon D, Ellis-Davies GCR, Frontera J, De Brito Van Velze M, Lourenco J, et al. GluN3A excitatory glycine receptors control adult cortical and amygdalar circuits. Neuron. 2022;110:2438–54.e8.
Hurley EP, Mukherjee B, Fang LZ, Barnes JR, Barron JC, Nafar F, et al. GluN3A and excitatory glycine receptors in the adult hippocampus. J Neurosci. 2024;44:e0401242024.
Otsu Y, Darcq E, Pietrajtis K, Matyas F, Schwartz E, Bessaih T, et al. Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science. 2019;366:250–4.
Grand T, Abi Gerges S, David M, Diana MA, Paoletti P. Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun. 2018;9:4769.
Zeng Y, Zheng YM, Zhang TT, Ye F, Zhan L, Kou ZW, et al. Identification of a subtype-selective allosteric inhibitor of GluN1/GluN3 NMDA receptors. Front Pharmacol. 2022;13:888308.
Zhu Z, Yi F, Epplin MP, Liu D, Summer SL, Mizu R, et al. Negative allosteric modulation of GluN1/GluN3 NMDA receptors. Neuropharmacology. 2020;176:108117.
Rogers M, Obergrussberger A, Kondratskyi A, Fertig N. Using automated patch clamp electrophysiology platforms in ion channel drug discovery: an industry perspective. Expert Opin Drug Discov. 2024;19:523–35.
Voldřich J, Matoušová M, Šmídková M, Mertlíková-Kaiserová H. Fluorescence-based HTS assays for ion channel modulation in drug discovery pipelines. ChemMedChem. 2024;19:e202400383.
Yu H, Li M, Wang W, Wang X. High throughput screening technologies for ion channels. Acta Pharmacol Sin. 2016;37:34–43.
Danel T, Łęski J, Podlewska S, Podolak IT. Docking-based generative approaches in the search for new drug candidates. Drug Discov Today. 2023;28:103439.
Vemula D, Jayasurya P, Sushmitha V, Kumar YN, Bhandari V. CADD, AI and ML in drug discovery: a comprehensive review. Eur J Pharm Sci. 2023;181:106324.
Wu Z, Chen S, Wang YH, Li FY, Xu HH, Li MX, et al. Current perspectives and trend of computer-aided drug design: a review and bibliometric analysis. Int J Surg. 2024;110:3848–78.
Bae H, Nam H. GraphATT-DTA: attention-based novel representation of interaction to predict drug-target binding affinity. Biomedicines. 2022;11:67.
Zhang S, Jiang MJ, Wang S, Wang XF, Wei ZQ, Li Z. SAG-DTA: prediction of drug-target affinity using self-attention graph network. Int J Mol Sci. 2021;22:8993.
Chen GX, He HH, Lv QJ, Zhao L, Chen CY-C. MMFA-DTA: multimodal feature attention fusion network for drug-target affinity prediction for drug repurposing against SARS-CoV-2. J Chem Theory Comput. 2024;20:8071–87.
Alqutaibi AY, Algabri RS, Alamri AS, Alhazmi LS, Almadani SM, Alturkistani AM, et al. Advancements of artificial intelligence algorithms in predicting dental implant prognosis from radiographic images: a systematic review. J Prosthet Dent. 2024;27:727–3.
Öztürk H, Özgür A, Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics. 2018;34:i821–29.
Han L, Kang L, Guo Q. ImageDTA: a simple model for drug–target binding affinity prediction. ACS Omega. 2024;9:28485–93.
Sirakanyan SN, Noravyan AS, Dzhagatspanyan IA, Nazaryan IM, Ovakimyan AA, Akopyan AG, et al. Synthesis and neurotropic activity of new derivatives of piperazino-substituted pyrano[3,4-c]pyridines. Pharm Chem J. 2013;46:591–4.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–8.
Yuan W, Chen G, Chen CY-C. FusionDTA: attention-based feature polymerizer and knowledge distillation for drug-target binding affinity prediction. Brief Bioinform. 2022;23:1–13.
Zhu X, Liu J, Zhang J, Yang ZH, Ynag F, Zhang XL. FingerDTA: a fingerprint-embedding framework for drug-target binding affinity prediction. Big Data Min Analytics. 2023;6:1–10.
Li Z, Ren P, Yang H, Zheng J, Bai F. TEFDTA: a transformer encoder and fingerprint representation combined prediction method for bonded and non-bonded drug-target affinities. Bioinformatics. 2024;40:1–8.
Forouzanfar F, Ahmadzadeh AM, Pourbagher-Shahri AM, Gorji A. Significance of NMDA receptor-targeting compounds in neuropsychological disorders: an in-depth review. Eur J Pharmacol. 2025;999:177690.
Öztürk H, Ozkirimli E, Özgür A. WideDTA: prediction of drug-target binding affinity. 2019;1–11. Preprint at https://doi.org/10.48550/arXiv.1902.04166.
Acknowledgements
This work was supported by the Dalian Science and Technology Innovation Fund Program (Grant ID: 2022JJ12GX017), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant ID: XDB0830403), the United Foundation for Medico-engineering Cooperation from Dalian Neusoft University of Information and the Second Hospital of Dalian Medical University (Grant ID: LH-JSRZ-202201), the Technology Innovation Project of Dalian Neusoft University of Information (Grant ID: TIFP202302) and the National Science and Technology Innovation 2030 Major Program (Grant ID: 2021ZD0200900). We are grateful for support from the Neusoft Research Institute of Dalian Neusoft University of Information.
Author information
Authors and Affiliations
Contributions
LH, LK, QG and ZBG designed the methodology and supervised the project. LH, XMZ, TYZ, and ZBY developed and implemented the virtual screening workflow. LH performed the formal analysis and initial data interpretation. XMZ, TYZ, and ZBY conducted additional data analyses and contributed to software deployment. LK supervised the data curation, provided critical resources, and validated the findings. ZBG and YZ designed the wet-lab experiments. YZ, SF, YSD and HYW conducted FDSS/μCell screening and whole-cell patch clamp recording. ZYQ performed homology modeling, binding pocket prediction, and molecular docking analyses. LH and YZ drafted the manuscript. LK, QG and ZBG reviewed and edited the manuscript. All authors approved the final manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, L., Zeng, Y., Qu, Zy. et al. Identification of small-molecule inhibitors for GluN1/GluN3A NMDA receptors via a multiscale CNN-based prediction model. Acta Pharmacol Sin 47, 32–40 (2026). https://doi.org/10.1038/s41401-025-01630-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01630-7
Keywords
This article is cited by
-
New advances in small molecule drugs targeting NMDA receptors
Acta Pharmacologica Sinica (2026)
-
AI-driven breakthroughs in ion channel drug discovery: the future is now
Acta Pharmacologica Sinica (2026)