Abstract
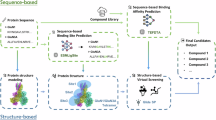
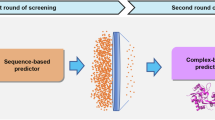
N-methyl-D-aspartate receptors (NMDARs) are calcium-permeable ionotropic glutamate receptors broadly expressed throughout the central nervous system, where they play crucial roles in neuronal development and synaptic plasticity. Among the various subtypes, the GluN1/GluN3A receptor represents a unique glycine-gated NMDAR with notably low calcium permeability. Despite its distinctive properties, GluN1/GluN3A remains understudied, particularly with respect to pharmacological tools development. This scarcity poses challenges for deeper investigation into its physiological functions and therapeutic relevance. In this study, we employed a hybrid virtual screening (VS) pipeline that integrates ligand-based and structure-based approaches for the efficient and precise identification of small-molecule candidates targeting GluN1/GluN3A. A large compound library comprising 18 million molecules was screened using an AI-enhanced multi-stage method. The initial phase utilized shape similarity ranking via ROCS-BART, followed by refinement with a graph neural network (GNN)-based drug-target interaction model to enhance docking accuracy. Functional validation using calcium flux (FDSS/μCell) identified two compounds with IC50 values below 10 μM. Of these, one candidate exhibited potent inhibitory activity with an IC50 of 5.31 ± 1.65 μM, which was further confirmed through manual patch-clamp recordings. These findings highlight an AI-enhanced VS workflow that achieves both efficiency and precision, providing a promising framework for exploring elusive targets such as GluN1/GluN3A.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol Rev. 2021;73:298–487.
Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400.
Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, et al. Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol. 2018;150:1081–105.
Ahmed H, Haider A, Ametamey SM. N-Methyl-D-Aspartate (NMDA) receptor modulators: a patent review (2015-present). Expert Opin Ther Pat. 2020;30:743–67.
Egunlusi AO, Joubert J. NMDA receptor antagonists: emerging insights into molecular mechanisms and clinical applications in neurological disorders. Pharmaceuticals. 2024;17:639
Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–8.
Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, et al. Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol. 2002;87:2052–63.
Pérez-Otaño I, Larsen RS, Wesseling JF. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci. 2016;17:623–35.
Marco S, Giralt A, Petrovic MM, Pouladi MA, Martínez-Turrillas R, Martínez-Hernández J, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med. 2013;19:1030–8.
Yuan T, Mameli M, O’Connor EC, Dey PN, Verpelli C, Sala C, et al. Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron. 2013;80:1025–38.
Mueller HT, Meador-Woodruff JH. NR3A NMDA receptor subunit mRNA expression in schizophrenia, depression and bipolar disorder. Schizophr Res. 2004;71:361–70.
Beesley S, Sullenberger T, Crotty K, Ailani R, D’Orio C, Evans K, et al. D-serine mitigates cell loss associated with temporal lobe epilepsy. Nat Commun. 2020;11:4966.
Pfisterer U, Petukhov V, Demharter S, Meichsner J, Thompson JJ, Batiuk MY, et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun. 2020;11:5038.
Grand T, Abi Gerges S, David M, Diana MA, Paoletti P. Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun. 2018;9:4769.
Kvist T, Greenwood JR, Hansen KB, Traynelis SF, Bräuner-Osborne H. Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors. Neuropharmacology. 2013;75:324–36.
Zhu Z, Yi F, Epplin MP, Liu D, Summer SL, Mizu R, et al. Negative allosteric modulation of GluN1/GluN3 NMDA receptors. Neuropharmacology. 2020;176:108117.
Zeng Y, Zheng Y, Zhang T, Ye F, Zhan L, Kou Z, et al. Identification of a subtype-selective allosteric inhibitor of GluN1/GluN3 NMDA receptors. Front Pharmacol. 2022;13:888308.
Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–53.
Bettini E, Sava A, Griffante C, Carignani C, Buson A, Capelli AM, et al. Identification and characterization of novel NMDA receptor antagonists selective for NR2A- over NR2B-containing receptors. J Pharmacol Exp Ther. 2010;335:636–44.
Yi F, Mou TC, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, et al. Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron. 2016;91:1316–29.
Hackos DH, Lupardus PJ, Grand T, Chen Y, Wang TM, Reynen P, et al. Positive allosteric modulators of GluN2A-containing NMDARs with distinct modes of action and impacts on circuit function. Neuron. 2016;89:983–99.
Vázquez J, López M, Gibert E, Herrero E, Luque FJ. Merging ligand-based and structure-based methods in drug discovery: an overview of combined virtual screening approaches. Molecules. 2020;25:4723.
Hawkins PC, Skillman AG, Nicholls A. Comparison of shape-matching and docking as virtual screening tools. J Med Chem. 2007;50:74–82.
Lewis M, Yinhan L, Goyal N, Ghazvininejad M, Mohamed A, Levy O, et al. BART: denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In: Proceedings of the 58th annual meeting of the association for computational linguistics. Association for Computational; 2020. pp. 7871–80. online.
Moon S, Hwang S-Y, Lim J, Kim WY. PIGNet2: a versatile deep learning-based protein–ligand interaction prediction model for binding affinity scoring and virtual screening. Digital Discov. 2024;3:287–99.
Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure. 2013;21:1788–99.
Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–91.
Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–52.
Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56.
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25.
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–90.
Bepari AK, Reza HM. Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation. PeerJ. 2021;9:e11261.
Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–9.
Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–91.
Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50.
Volgraf M, Sellers BD, Jiang Y, Wu G, Ly CQ, Villemure E, et al. Discovery of GluN2A-selective NMDA receptor positive allosteric modulators (PAMs): tuning deactivation kinetics via structure-based design. J Med Chem. 2016;59:2760–79.
Irwin R, Dimitriadis S, He J, Bjerrum EJ. Chemformer: a pre-trained transformer for computational chemistry. Mach Learn Sci Technol. 2022;3:015022.
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49.
Shen C, Zhang X, Hsieh CY, Deng Y, Wang D, Xu L, et al. A generalized protein-ligand scoring framework with balanced scoring, docking, ranking and screening powers. Chem Sci. 2023;14:8129–46.
Bossi S, Pizzamiglio L, Paoletti P. Excitatory GluN1/GluN3A glycine receptors (eGlyRs) in brain signaling. Trends Neurosci. 2023;46:667–81.
Villemure E, Volgraf M, Jiang Y, Wu G, Ly CQ, Yuen PW, et al. GluN2A-selective pyridopyrimidinone series of NMDAR positive allosteric modulators with an improved in vivo profile. ACS Med Chem Lett. 2016;8:84–9.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Bondi A. van der Waals volumes and radii. J Phys Chem. 1964;68:441–51.
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant ID: XDB0830403) and the National Science and Technology Innovation 2030 Major Program (Grant ID: 2021ZD0200900). The authors gratefully acknowledge Professor Mao-lin Wang from the First Affiliated Hospital of Shantou University Medical College for his invaluable technical support and assistance with the OpenEye software.
Author information
Authors and Affiliations
Contributions
YK and ZBG designed the study. YK, YSJ, SFH, BCZ, SWL, and CL designed the virtual screening strategy. BCZ built the homology model and analyzed the structure-activity relationship. SWL, SFH and YK constructed the ROCS-BART model and deployed PIGNet2. YSJ calculated the virtual screening scores. YSJ and BCZ analyzed the virtual screening results and selected the candidate compounds. ZBG and YZ designed wet-lab experiments. YZ, HCW and AYW conducted FDSS/μCell screening and whole-cell patch clamp recording. YSJ, SFH and YZ wrote the manuscript. All authors reviewed the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ji, Ys., Zeng, Y., Hu, Sf. et al. AI-enhanced virtual screening approach to hit identification for GluN1/GluN3A NMDA receptor. Acta Pharmacol Sin 47, 41–52 (2026). https://doi.org/10.1038/s41401-025-01644-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01644-1
Keywords
This article is cited by
-
AI-driven breakthroughs in ion channel drug discovery: the future is now
Acta Pharmacologica Sinica (2026)
-
New advances in small molecule drugs targeting NMDA receptors
Acta Pharmacologica Sinica (2026)