Abstract
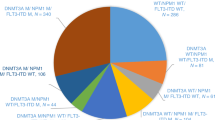
Acute myeloid leukemia (AML) includes genetically defined subsets. In allogeneic hematopoietic cell transplantation (allo-HCT), the frequency and prognosis of gene-gene interactions may differ from those of patients treated with chemotherapy alone. In this study, adult patients (N = 952) with AML allografted between 2015 and 2023, with available next generation sequencing (NGS) at diagnosis were included. Most frequent mutations were DNMT3A (24%), FLT3-ITD (21%), NPM1 (21%), RUNX1 (16%), NRAS (16%), TET2 (14%), and IDH2 (12%). Multiple correspondence analysis identified distinct groups of co-occurring mutations. Outcome analysis was performed on 646 AML patients allografted in first complete remission (CR1). Six non-overlapping groups were constructed: 1) TP53 mutation (N = 47); 2) NPM1 mutation (N = 129); 3) FLT3-ITD and/or DNMT3A mutation (N = 128); 4) SRSF2 and/or ASXL1 and/or RUNX1 mutation (SAR group) (N = 132); 5) IDH1 and/or IDH2 and/or TET2 mutation (N = 43); and 6) all ten genes unmutated (N = 167). In multivariable analysis, TP53 mutation, adverse karyotype, and age negatively affected leukemia-free survival (LFS) and overall survival (OS). OS was additionally negatively affected when the ten genes were unmutated. Notably, outcomes were excellent for SAR mutations (2-year LFS 76%, OS 84%), indicating allo-HCT in CR1 can overcome their adverse risk at diagnosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data analyzed in this study were provided and approved by the ALWP of the EBMT. All relevant data supporting the findings of this study are available within the Article and the Supplementary Material. Requests for access to EBMT study data from qualified external researchers will be reviewed by the relevant working party in accordance with EBMT’s data-sharing policies. This process ensures the protection of patient privacy, maintains data security and integrity, and promotes scientific and medical innovation. Additional details on data-sharing criteria and the request process can be obtained by contacting ebmt.do-paris@ebmt.org. Individual patient-level data will not be shared.
References
DiNardo CD, Erba HP, Freeman SD, Wei AH. Acute myeloid leukaemia. Lancet. 2023;401:2073–86. https://doi.org/10.1016/s0140-6736(23)00108-3.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. https://doi.org/10.1056/NEJMoa1516192.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. https://doi.org/10.1182/blood-2009-11-254441.
Romer-Seibert JS, Meyer SE. Genetic heterogeneity and clonal evolution in acute myeloid leukemia. Current Opin Hematol. 2021;28:64–70. https://doi.org/10.1097/moh.0000000000000626.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77. https://doi.org/10.1182/blood.2022016867.
Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood. 2016;127:62–70. https://doi.org/10.1182/blood-2015-07-604546.
Bacigalupo A, Ballen K, Rizzo D, Giralt S, Lazarus H, Ho V, et al. Defining the intensity of conditioning regimens: working definitions. Biology Blood Marrow Transplant. 2009;15:1628–33. https://doi.org/10.1016/j.bbmt.2009.07.004.
Spyridonidis A, Labopin M, Savani BN, Niittyvuopio R, Blaise D, Craddock C, et al. Redefining and measuring transplant conditioning intensity in current era: a study in acute myeloid leukemia patients. Bone Marrow Transpl. 2020;55:1114–25. https://doi.org/10.1038/s41409-020-0803-y.
Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304. https://doi.org/10.1097/00007890-197410000-00001.
Luskin MR, Carroll M, Lieberman D, Morrissette JJD, Zhao J, Crisalli L, et al. Clinical utility of next-generation sequencing for oncogenic mutations in patients with acute myeloid leukemia undergoing allogeneic stem cell transplantation. Biology Blood Marrow Transplant. 2016;22:1961–7. https://doi.org/10.1016/j.bbmt.2016.07.018.
Yoshizato T, Nannya Y, Atsuta Y, Shiozawa Y, Iijima-Yamashita Y, Yoshida K, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129:2347–58. https://doi.org/10.1182/blood-2016-12-754796.
Ahn JS, Kim HJ, Kim YK, Lee SS, Jung SH, Yang DH, et al. DNMT3A R882 mutation with FLT3-ITD positivity is an extremely poor prognostic factor in patients with normal-karyotype acute myeloid leukemia after allogeneic hematopoietic cell transplantation. Biology Blood Marrow Transplant. 2016;22:61–70. https://doi.org/10.1016/j.bbmt.2015.07.030.
Mohty R, Bazarbachi AH, Labopin M, Esteve J, Kröger N, Cornelissen JJ, et al. Isocitrate dehydrogenase (IDH) 1 and 2 mutations predict better outcome in patients with acute myeloid leukemia undergoing allogeneic hematopoietic cell transplantation: a study of the ALWP of the EBMT. Bone Marrow Transpl. 2024;59:1534–41. https://doi.org/10.1038/s41409-024-02384-2.
Abou Dalle I, Galimard J-E, Poire X, Huynh A, Wagner Drouet E, Burns D, et al. The impact of DNMT3A mutation on survival of AML patients receiving allogeneic hematopoietic cell transplantation in first remission depends on the karyotype and co-occurring mutations: on Behalf of the EBMT Acute Leukemia Working Party. Blood. 2023;142:658. https://doi.org/10.1182/blood-2023-179531.
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. https://doi.org/10.1056/NEJMoa1112304.
Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–8. https://doi.org/10.1038/leu.2016.126.
Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol. 2012;30:3109–18. https://doi.org/10.1200/jco.2011.40.6652.
Paschka P, Schlenk RF, Gaidzik VI, Herzig JK, Aulitzky T, Bullinger L, et al. ASXL1 mutations in younger adult patients with acute myeloid leukemia: a study by the German-Austrian Acute Myeloid Leukemia Study Group. Haematologica. 2015;100:324–30. https://doi.org/10.3324/haematol.2014.114157.
Weinberg OK, Siddon A, Madanat YF, Gagan J, Arber DA, Dal Cin P, et al. TP53 mutation defines a unique subgroup within complex karyotype de novo and therapy-related MDS/AML. Blood Adv. 2022;6:2847–53. https://doi.org/10.1182/bloodadvances.2021006239.
Middeke JM, Herold S, Rücker-Braun E, Berdel WE, Stelljes M, Kaufmann M, et al. TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Brit J Haematol. 2016;172:914–22. https://doi.org/10.1111/bjh.13912.
Zhao D, Zarif M, Zhou Q, Capo-Chichi JM, Schuh A, Minden MD, et al. TP53 mutations in AML patients are associated with dismal clinical outcome irrespective of frontline induction regimen and allogeneic hematopoietic cell transplantation. Cancers. 2023;15:3210 https://doi.org/10.3390/cancers15123210.
Murdock HM, Kim HT, Denlinger N, Vachhani P, Hambley B, Manning BS, et al. Impact of diagnostic genetics on remission MRD and transplantation outcomes in older patients with AML. Blood. 2022;139:3546–57. https://doi.org/10.1182/blood.2021014520.
Nawas MT, Kosuri S. Utility or futility? A contemporary approach to allogeneic hematopoietic cell transplantation for TP53-mutated MDS/AML. Blood Adv. 2024;8:553–61. https://doi.org/10.1182/bloodadvances.2023010417.
Shahzad M, Tariq E, Chaudhary SG, Anwar I, Iqbal Q, Fatima H, et al. Outcomes with allogeneic hematopoietic stem cell transplantation in TP53-mutated acute myeloid leukemia: a systematic review and meta-analysis. Leukemia lymphoma. 2022;63:3409–17. https://doi.org/10.1080/10428194.2022.2123228.
Loke J, Labopin M, Craddock C, Cornelissen JJ, Labussière-Wallet H, Wagner-Drouet EM, et al. Additional cytogenetic features determine outcome in patients allografted for TP53 mutant acute myeloid leukemia. Cancer. 2022;128:2922–31. https://doi.org/10.1002/cncr.34268.
Nagler A, Labopin M, Salmenniemi U, Wu D, Blaise D, Rambaldi A, et al. Trends in allogeneic transplantation for favorable risk acute myeloid leukemia in first remission: a longitudinal study of >15 years from the ALWP of the EBMT. Bone Marrow Transpl. 2024;59:1563–76. https://doi.org/10.1038/s41409-024-02379-z.
Othman J, Potter N, Ivey A, Tazi Y, Papaemmanuil E, Jovanovic J, et al. Molecular, clinical, and therapeutic determinants of outcome in NPM1-mutated AML. Blood. 2024;144:714–28. https://doi.org/10.1182/blood.2024024310.
Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med. 2017;377:454–64. https://doi.org/10.1056/NEJMoa1614359.
Bazarbachi A, Labopin M, Battipaglia G, Djabali A, Forcade E, Arcese W, et al. Allogeneic stem cell transplantation for FLT3-mutated acute myeloid leukemia: in vivo T-cell depletion and posttransplant sorafenib maintenance improve survival. A retrospective acute leukemia Working Party-European Society for Blood and Marrow Transplant Study. Clin Hematol Int. 2019;1:58–74. https://doi.org/10.2991/chi.d.190310.001.
Burchert A, Bug G, Fritz LV, Finke J, Stelljes M, Röllig C, et al. Sorafenib maintenance after allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia with FLT3-internal tandem duplication mutation (SORMAIN). J Clin Oncol. 2020;38:2993–3002. https://doi.org/10.1200/jco.19.03345.
Abou Dalle I, El Cheikh J, Bazarbachi A. Pharmacologic strategies for post-transplant maintenance in acute myeloid leukemia: it is time to consider!. Cancers. 2022;14:1490 https://doi.org/10.3390/cancers14061490.
Stone RM, Yin J, Mandrekar SJ, Benner A, Saadati M, Galinsky IA, et al. 10 year follow-up of CALGB 10603/ratify: midostaurin versus placebo plus intensive chemotherapy in newly diagnosed FLT3 mutant acute myeloid leukemia patients aged 18-60 years. Blood. 2024;144:218. https://doi.org/10.1182/blood-2024-201058.
Bazarbachi A, Labopin M, Gedde-Dahl T, Remenyi P, Forcade E, Kröger N, et al. Improved posttransplant outcomes in recent years for AML patients with FLT3-ITD and wild-type NPM1: a report from the EBMT Acute Leukemia Working Party. Clinical Cancer Res. 2023;29:4441–8. https://doi.org/10.1158/1078-0432.ccr-23-0954.
Pandya BJ, Burns LJ, Wang T, Xie B, Touya M, Spalding J, et al. Clinical outcomes and treatment patterns in adults with FLT3-ITD(mut+) acute myeloid leukemia undergoing allogeneic hemopoietic cell transplantation in the United States and Canada. Transplant Cell Ther. 2024;30:683.e681–683.e613. https://doi.org/10.1016/j.jtct.2024.04.016.
Gardin C, Pautas C, Fournier E, Itzykson R, Lemasle E, Bourhis JH, et al. Added prognostic value of secondary AML-like gene mutations in ELN intermediate-risk older AML: ALFA-1200 study results. Blood Adv. 2020;4:1942–9. https://doi.org/10.1182/bloodadvances.2019001349.
Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76. https://doi.org/10.1182/blood-2014-11-610543.
van der Werf I, Wojtuszkiewicz A, Meggendorfer M, Hutter S, Baer C, Heymans M, et al. Splicing factor gene mutations in acute myeloid leukemia offer additive value if incorporated in current risk classification. Blood Adv. 2021;5:3254–65. https://doi.org/10.1182/bloodadvances.2021004556.
Alawieh D, Cysique-Foinlan L, Willekens C, Renneville A. RAS mutations in myeloid malignancies: revisiting old questions with novel insights and therapeutic perspectives. Blood Cancer J. 2024;14:72. https://doi.org/10.1038/s41408-024-01054-2.
Fobare S, Kohlschmidt J, Ozer HG, Mrózek K, Nicolet D, Mims AS, et al. Molecular, clinical, and prognostic implications of PTPN11 mutations in acute myeloid leukemia. Blood Adv. 2022;6:1371–80. https://doi.org/10.1182/bloodadvances.2021006242.
Heuser M, Gabdoulline R, Löffeld P, Dobbernack V, Kreimeyer H, Pankratz M, et al. Individual outcome prediction for myelodysplastic syndrome (MDS) and secondary acute myeloid leukemia from MDS after allogeneic hematopoietic cell transplantation. Ann Hematol. 2017;96:1361–72. https://doi.org/10.1007/s00277-017-3027-5.
Acknowledgements
Patients and caregivers, EBMT participating centers, the ALWP statistical and data management team, particularly Aleksandra (Sasha) Gavrilina, as well as the leadership team. This was presented in part during the American Society of Hematology meeting in San Diego, December 2023.
Author information
Authors and Affiliations
Contributions
ABa proposed the study, interpreted the data, and wrote the manuscript. J-EG and MMo participated in study design, data interpretation, and manuscript editing. J-EG performed the statistical analysis. IAD and MLa contributed to data interpretation and manuscript revision. JS, HH, JM, CS, BL, LG, JMa, MI-R, AK, M-PG-H, GB, J-MR, AG, CSc, MK, XP, PC, MJC, FB, CC, EB, AN, and FC collected and updated patient data, and critically reviewed and approved the manuscript. All authors reviewed the final version, approved the data presented, and agreed with the conclusions of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This is a retrospective, registry-based, multicenter study utilizing patient data collected and approved by the Acute Leukemia Working Party (ALWP) of the EBMT. The EBMT is a collaborative network comprising more than 600 transplant centers that are required to report all consecutive HCTs and subsequent follow-ups on an annual basis. Routine audits are performed to ensure data accuracy and completeness. Since January 2003, all participating transplant centers have been required to obtain written informed consent from patients prior to data registration with the EBMT, in accordance with the ethical principles outlined in the Declaration of Helsinki (1975).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bazarbachi, A., Galimard, JE., Abou Dalle, I. et al. Frequency and impact of somatic co-occurring mutations on post-transplant outcomes in acute myeloid leukemia: a multicenter registry analysis on behalf of the EBMT ALWP. Bone Marrow Transplant (2025). https://doi.org/10.1038/s41409-025-02770-4
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41409-025-02770-4