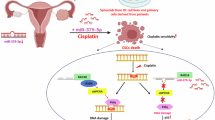
Abstract
Background
Cancer stem-like cells (CSLCs) drive tumour progression and chemoresistance. The concerted efforts of EXO1 and TLS polymerases safeguard DNA integrity against chemotherapeutic drugs. In absence of potential drug targets, non-small cell lung carcinoma (NSCLC) patients have few therapeutic options. In current scenario, microRNAs offer a potential avenue for eradicating CSLCs.
Methods
EXO1 downregulation impact on CSLCs expansion was assessed via flow cytometry. Co-localisation of EXO1, Polη and Polι was validated through co-immunoprecipitation and confocal-imaging. The effects of co-downregulation of Polη and Polι on CSLC survival, repair synthesis, and mutagenesis were evaluated using flow cytometry and immunohistochemistry in cell lines and xenografts. MicroRNA targeting EXO1 was studied for its role in CSLCs regulation.
Results
EXO1 downregulation in NSCLC CSLCs induces DNA lesions, triggering apoptosis and enhances cisplatin sensitivity. It collaborates with Polη and Polι in DNA repair, contributing to cisplatin resistance in CSLCs. Absence of Polη and Polι impairs repair and reduces cisplatin-induced mutagenesis. Co-downregulation of Polη and Polι in xenografts reduces tumour proliferation significantly. MiR-3163 overexpression sensitises CSLCs to cisplatin via targeting EXO1/Polη/Polι axis, as shown in mechanistic studies.
Conclusion
This study unveils a novel regulatory pathway involving EXO1/Polη/Polι axis and miR-3163, providing insights into CSLCs regulation in NSCLC.

EXO1/Polη/Polι axis targeted by miR-3163, resulting in the inhibition of cell growth and induction of apoptosis in NSCLC CSLCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Franco S, Szczesna K, Iliou MS, Al-Qahtani M, Mobasheri A, Kobolák J, et al. In vitro models of cancer stem cells and clinical applications. BMC Cancer. 2016;16:738.
Wang C, Kulkarni P, Salgia R. Combined checkpoint inhibition and chemotherapy: new era of 1st-line treatment for non-small-cell lung cancer. Mol Ther Oncolytics. 2019;13:1–6.
Kamal A, Wessel M, Wyant T, Zhang Q, Alteri R, Lubejko B, et al. Lung cancer—non-small cell: statistics. 2023. https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics.
Zhou J, Wang Y, Wang Y, Yin X, He Y, Chen L, et al. FOXM1 modulates cisplatin sensitivity by regulating EXO1 in ovarian cancer. PLoS ONE. 2014;9:e96989.
Zhou C-S, Feng M-T, Chen X, Gao Y, Chen L, Li L-D, et al. Exonuclease 1 (EXO1) is a potential prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Onco Targets Ther. 2021;14:1033–48.
Liu J, Zhang J. Elevated EXO1 expression is associated with breast carcinogenesis and poor prognosis. Ann Transl Med. 2021;9:135.
Rein K, Yanez DA, Terré B, Palenzuela L, Aivio S, Wei K, et al. EXO1 is critical for embryogenesis and the DNA damage response in mice with a hypomorphic Nbs1 allele. Nucleic Acids Res. 2015;43:7371–87.
He D, Li T, Sheng M, Yang B. Exonuclease 1 (Exo1) participates in mammalian non-homologous end joining and contributes to drug resistance in ovarian cancer. Med Sci Monit. 2020;26:e918751.
Mao P, Wu S, Fan Y. Upregulation of exonuclease 1 caused by homology-dependent repair confers cisplatin resistance to gastric cancer cells. Can J Physiol Pharm. 2022;100:903–14.
Sertic S, Mollica A, Campus I, Roma S, Tumini E, Aguilera A, et al. Coordinated activity of Y family TLS polymerases and EXO1 protects non-S phase cells from UV-induced cytotoxic lesions. Mol Cell. 2018;70:34–47.e4.
Pilzecker B, Jacobs H. Mutating for good: DNA damage responses during somatic hypermutation. Front Immunol. 2019;10:438.
Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363.
Srivastava A, Han C, Zhao R, Cui T, Dai Y, Mao H, et al. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc Natl Acad Sci USA. 2015;112:4411–6.
Li L, Tian H, Cheng C, Li S, Ming L, Qi L. SiRNA of DNA polymerase iota inhibits the migration and invasion in the lung cancer cell A549. Acta Biochim Biophys Sin. 2018;50:929–33.
Zhou HM, Zhang JG, Zhang X, Li Q. Targeting cancer stem cells for reversing therapy resistance: mechanism, signaling, and prospective agents. Signal Transduct Target Ther. 2021;6:62.
Bartel D. MicroRNA target recognition and regulatory functions. Cell. 2009;136:215–33.
Qiu P, Guo Q, Yao Q, Chen J, Lin J. Hsa-mir-3163 and CCNB1 may be potential biomarkers and therapeutic targets for androgen receptor positive triple-negative breast cancer. PLoS ONE. 2021;16:e0254283.
Delgir S, Ilkhani K, Safi A, Rahmati Y, Montazari V, Zaynali-Khasraghi Z, et al. The expression of miR-513c and miR-3163 was downregulated in tumor tissues compared with normal adjacent tissue of patients with breast cancer. BMC Med Genomics. 2021;14:180.
Ren H, Li Z, Tang Z, Li J, Lang X. Long noncoding MAGI2-AS3 promotes colorectal cancer progression through regulating miR-3163/TMEM106B axis. J Cell Physiol. 2020;235:4824–33.
Liu D, Zhang H, Cong J, Cui M, Ma M, Zhang F, et al. H3K27 acetylation-induced lncRNA EIF3J-AS1 improved proliferation and impeded apoptosis of colorectal cancer through miR-3163/YAP1 axis. J Cell Biochem. 2020;121:1923–33.
Su L, Han D, Wu J, Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour Biol. 2015;37:3925–31.
Zhu W, Yamasaki H, Mironov N. Frequency of HPRT gene mutations induced by N-methyl-N X-nitro-N-nitrosoguanidine corresponds to replication error phenotypes of cell lines. Mutat Res. 1998;398:93–99.
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27.
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46.
Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ, Gimotty PA, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898–911.
Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, et al. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–45.
Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–65.
Wang Z, Chao Z, Wang Q, Zou F, Song T, Xu L, et al. EXO1/P53/SREBP1 axis-regulated lipid metabolism promotes prostate cancer progression. J Transl Med. 2024;22:104.
Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–71.
Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature. 2009;460:1132–5.
Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014;74:1554–65.
Novikov NM, Zolotaryova SY, Gautreau AM, Denisov EV. Mutational drivers of cancer cell migration and invasion. Br J Cancer. 2021;124:102–14.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–31.
Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239.
Ren H, Li Z, Tang Z, Li J, Lang X. Long noncoding MAGI2-AS3 promotes colorectal cancer progression through regulating miR-3163/TMEM106B axis. JCP. 2019;235:4824–33.
Liu D, Zhang H, Cong J, Cui M, Ma M, Zhang F, et al. H3K27 acetylation-induced lncRNA EIF3J-AS1 improved proliferation and impeded apoptosis of colorectal cancer through miR-3163/YAP1 axis. J Cell Biochem. 2019;121:1923–33.
Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase η in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res. 2006;4:257–65.
Ball LG, Hanna MD, Lambrecht AD, Mitchell BA, Ziola B, Cobb JA, et al. The Mre11-Rad50-Xrs2 complex is required for yeast DNA post replication repair. PLoS ONE. 2014;9:e109292.
Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500.
Kannouche P, Lehmann A. Localization of Y‐family polymerases and the DNA polymerase switch in mammalian cells. Methods Enzymol. 2006;408:407–15.
Kannouche P, Henestrosa A, Coull B, Vidal A, Gray C, Zicha D, et al. Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2002;21:6246–56.
Jia M, Wei Z, Liu P, Zhao X. Silencing of ABCG2 by microRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. J Korean Med Sci. 2016;31:836–42.
Acknowledgements
TM and DS are recipients of Senior Research Fellowship from CSIR. Also, the authors are thankful to Prof. Qi-En Wang and Prof. Ramesh Ganju from The Ohio State University for providing the cell lines.
Funding
The study was supported by ICMR, New Delhi (File no. 2020-4993/SCR/ADHOC-BMS).
Author information
Authors and Affiliations
Contributions
Tanima Mandal: conceptualisation, methodology, investigation, formal analysis, writing—original draft; Devendra Shukla: conceptualisation, methodology, investigation, formal analysis, writing—original draft; Md Maqsood Ahamad Khan: investigation; Senthil Kumar Ganesan: formal analysis; Amit Kumar Srivastava: conceptualisation, methodology, resources, investigation, formal analysis, validation, writing—reviewing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal protocols were approved by the CSIR-IICB-Animal ethics committee (IICB/AEC/Meeting/July/2020/1). All methods are reported following the ARRIVE guidelines (https://arriveguidelines.org) for animal experiment reporting, with ethics approval.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mandal, T., Shukla, D., Khan, M.M.A. et al. The EXO1/Polη/Polι axis as a promising target for miR-3163-mediated attenuation of cancer stem-like cells in non-small cell lung carcinoma. Br J Cancer 131, 1668–1682 (2024). https://doi.org/10.1038/s41416-024-02840-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41416-024-02840-2
This article is cited by
-
Targeting epigenetic regulators as a promising avenue to overcome cancer therapy resistance
Signal Transduction and Targeted Therapy (2025)
-
The role of extracellular vesicles in the communication between endometrial cancer cells and tumour-associated macrophages: a review
Journal of Cancer Research and Clinical Oncology (2025)