Abstract
Background
m1A, a prevalent RNA modification found in various RNA species, has recently been reported to modulate cancer progression. However, its effects on neuroblastoma remain uninvestigated.
Methods
The PCAT database was utilized to analyze the mRNA levels and survival probabilities of m1A regulator genes (TRMT6, TRMT61A, ALKBH1, and ALKBH3) in neuroblastoma patients. Silencing and recovery of TRMT6 were employed to investigate its role in neuroblastoma in vitro and in vivo. m1A-seq and RIP-qPCR were performed to identify and confirm the downstream targets of TRMT6. Additionally, Actinomycin D treatment was administered to assess mRNA stability.
Results
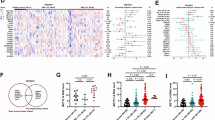
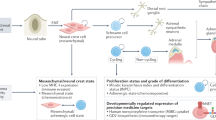
m1A transmethylase TRMT6 expression was significantly elevated in high-risk and late-stage neuroblastoma patients. Functionally, TRMT6 promotes the malignancy of neuroblastoma cells in vitro and promotes tumor growth and metastasis in vivo. Mechanistically, TRMT6 reduces SST mRNA levels by inhibiting its stability in an m1A-YTHDF2-dependent manner, thereby promoting the development of neuroblastoma. Furthermore, SST analog octreotide suppresses neuroblastoma cell malignancy, tumor growth, and metastasis.
Conclusions
TRMT6 mediates m1A modification of SST to promote neuroblastoma progression, suggesting that targeting TRMT6 may be a novel potential therapeutic approach for treating neuroblastoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data of the m1A-seq in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE297545. All data presented in this study are the available from the corresponding author upon request.
References
Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20.
Smith EI, Haase GM, Seeger RC, Brodeur GM. A surgical perspective on the current staging in neuroblastoma-the International Neuroblastoma Staging System proposal. J Pediatr Surg. 1989;24:386–90.
Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–97.
Brodeur GM. Spontaneous regression of neuroblastoma. Cell Tissue Res. 2018;372:277–86.
Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F, et al. Molecular targeting therapies for neuroblastoma: progress and challenges. Med Res Rev. 2021;41:961–1021.
Qiu B, Matthay KK. Advancing therapy for neuroblastoma. Nat Rev Clin Oncol. 2022;19:515–33.
Fetahu IS, Taschner-Mandl S. Neuroblastoma and the epigenome. Cancer Metastasis Rev. 2021;40:173–89.
Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23.
Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129.
Jin H, Huo C, Zhou T, Xie S. m(1)A RNA modification in gene expression regulation. Genes. 2022;13:910.
Li J, Zhang H, Wang H. N(1)-methyladenosine modification in cancer biology: current status and future perspectives. Comput Struct Biotechnol J. 2022;20:6578–85.
Zhang C, Jia G. Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genom Proteom Bioinform. 2018;16:155–61.
Oerum S, Dégut C, Barraud P, Tisné C. m1A post-transcriptional modification in tRNAs. Biomolecules. 2017;7:20.
Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251–5.
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–6.
Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–90.
Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017;68:993–1005.e1009.
Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, et al. Mitochondrial 16S rRNA Is Methylated by tRNA methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14:e1002557.
Kuang W, Jin H, Yang F, Chen X, Liu J, Li T, et al. ALKBH3-dependent m(1)A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022;8:25.
Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:816–828.e816.
Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12:6314.
Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, et al. tRNA-m(1)A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. 2022;23:1433–44.
Jiang C, Tian Y, Xu C, Zhang H, Gu L. Landscape of N1-methyladenosin (m1A) modification pattern in colorectal cancer. Cancer Rep. 2024;7:e1965.
Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R, et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun. 2022;13:2165.
Gu X, Zhuang A, Yu J, Yang L, Ge S, Ruan J, et al. Histone lactylation-boosted ALKBH3 potentiates tumor progression and diminished promyelocytic leukemia protein nuclear condensates by m1A demethylation of SP100A. Nucleic Acids Res. 2024;52:2273–89.
Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–62.
Monshaugen I, Luna L, Rhodes J, Kristiansen FIS, Lång A, Bøe SO, et al. Depletion of the m1A writer TRMT6/TRMT61A reduces proliferation and resistance against cellular stress in bladder cancer. Front Oncol. 2023;13:1334112.
Wang Y, Huang Q, Deng T, Li BH, Ren XQ. Clinical significance of TRMT6 in hepatocellular carcinoma: a bioinformatics-based study. Med Sci Monit. 2019;25:3894–901.
Ye Y, Liu M, Wu F, Ou S, Wang W, Fei J, et al. TRMT6 promotes hepatocellular carcinoma progression through the PI3K/AKT signaling pathway. Eur J Med Res. 2023;28:48.
Wang B, Niu L, Wang Z, Zhao Z. RNA m1A methyltransferase TRMT6 predicts poorer prognosis and promotes malignant behavior in glioma. Front Mol Biosci. 2021;8:692130.
Chang X, Zhu J, Hua RX, Deng C, Zhang J, Cheng J, et al. TRMT6 gene rs236110 C > A polymorphism increases the risk of Wilms tumor. Gene. 2023;882:147646.
Miao L, Zhuo Z, Tang J, Huang X, Liu J, Wang HY, et al. FABP4 deactivates NF-κB-IL1α pathway by ubiquitinating ATPB in tumor-associated macrophages and promotes neuroblastoma progression. Clin Transl Med. 2021;11:e395.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y, et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–81.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–7.
Seo KW, Kleiner RE. YTHDF2 recognition of N(1)-methyladenosine (m(1)A)-modified RNA is associated with transcript destabilization. ACS Chem Biol. 2020;15:132–9.
Zheng Q, Gan H, Yang F, Yao Y, Hao F, Hong L, et al. Cytoplasmic m(1)A reader YTHDF3 inhibits trophoblast invasion by downregulation of m(1)A-methylated IGF1R. Cell Discov. 2020;6:12.
Guan Q, Lin H, Hua W, Lin L, Liu J, Deng L, et al. Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression. Chin J Cancer Res. 2023;35:140–62.
Sun L, Coy DH. Somatostatin and its analogs. Curr Drug Targets. 2016;17:529–37.
Reubi JC, Laissue JA. Multiple actions of somatostatin in neoplastic disease. Trends Pharm Sci. 1995;16:110–5.
Cattaneo MG, Amoroso D, Gussoni G, Sanguini AM, Vicentini LM. A somatostatin analogue inhibits MAP kinase activation and cell proliferation in human neuroblastoma and in human small cell lung carcinoma cell lines. FEBS Lett. 1996;397:164–8.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20.
Zeng H, Li M, Liu J, Zhu J, Cheng J, Li Y, et al. YTHDF2 gene rs3738067 A>G polymorphism decreases neuroblastoma risk in Chinese children: evidence from an eight-center case-control study. Front Med. 2021;8:797195.
Pomaville MM, He C. Advances in targeting RNA modifications for anticancer therapy. Trends Cancer. 2023;9:528–42.
Li M, Ye J, Xia Y, Li M, Li G, Hu X, et al. METTL3 mediates chemoresistance by enhancing AML homing and engraftment via ITGA4. Leukemia. 2022;36:2586–95.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e123.
Cattaneo MG, Scita G, Vicentini LM. Somatostatin inhibits PDGF-stimulated Ras activation in human neuroblastoma cells. FEBS Lett. 1999;459:64–68.
Mercadante S, Porzio G. Octreotide for malignant bowel obstruction: twenty years after. Crit Rev Oncol Hematol. 2012;83:388–92.
Graillon T, Romano D, Defilles C, Saveanu A, Mohamed A, Figarella-Branger D, et al. Octreotide therapy in meningiomas: in vitro study, clinical correlation, and literature review. J Neurosurg. 2017;127:660–9.
Watson SA, Durrant LG, Morris DL. The effect of the E2 prostaglandin enprostil, and the somatostatin analogue SMS 201 995, on the growth of a human gastric cell line, MKN45G. Int J Cancer. 1990;45:90–94.
Dy DY, Whitehead RH, Morris DL. SMS 201.995 inhibits in vitro and in vivo growth of human colon cancer. Cancer Res. 1992;52:917–23.
Borgström P, Hassan M, Wassberg E, Refai E, Jonsson C, Larsson SA, et al. The somatostatin analogue octreotide inhibits neuroblastoma growth in vivo. Pediatr Res. 1999;46:328–32.
Funding
This work was supported by the National Natural Science Foundation of China (32300473,82002636, 82173593), Guangdong Basic and Applied Basic Research Foundation (2023A1515220053), the Science, Technology and Innovation Commission of Shenzhen (JCYJ20220531093213030).
Author information
Authors and Affiliations
Contributions
Xinxin Zhang, Zhenjian Zhuo, and Jing He designed this project. Xinxin Zhang, Huimin Yin, Liping Chen, and Mengzhen Zhang completed the in vitro experiments. Xinxin Zhang, Huiran Lin, Huimin Yin, Yufeng Han, and Hongxia Chen conducted the in vivo experiments. Jing He collected NB samples and related clinical data. Xinxin Zhang provided the data analysis. Xinxin Zhang, Zhenjian Zhuo, and Jing He wrote the paper. All authors approved this version to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures involving neuroblastoma patients in this study were conducted following the standards of the Ethics Committee of Guangzhou Women and Children’s Medical Center (Approval number: 2023-120A01). Written consent was signed by patients’ guardians prior to sample collection.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Lin, H., Yin, H. et al. m1A methylase TRMT6 promotes neuroblastoma development by demethylating SST mRNA in an m1A/YTHDF2-dependent manner. Br J Cancer 133, 1354–1364 (2025). https://doi.org/10.1038/s41416-025-03152-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41416-025-03152-9