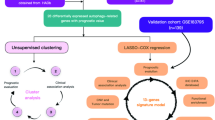
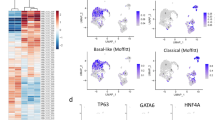
Abstract
Background
Prognostication for pancreatic ductal adenocarcinoma (PDAC) using histologic images is difficult due to tumor heterogeneity. We developed an artificial intelligence (AI) model to predict postoperative recurrence using histologic image patches.
Methods
We included 591 patients with resected PDAC to train an AI model for recurrence prediction at 12 or 24 months and validated it using external cohorts (n = 302 in total). Image patches from hematoxylin and eosin-stained slides were clustered via uniform manifold approximation and projection (UMAP) and used to train a random forest model. Predictive performance was evaluated using area under the receiver operating characteristic curve (AUC). Gene expression analysis was conducted to characterise survival-related clusters.
Results
Seventeen patch clusters were identified. Two were linked to high recurrence risk, and one to low risk. In external validation, the model achieved an AUC of up to 0.792. The random forest score independently predicted recurrence. Greater heterogeneity in patch composition correlated with shorter time to recurrence (P < 0.01). High-risk clusters showed elevated CSF3R expression; the low-risk cluster showed increased IGFBP3 expression.
Conclusions
Our AI model, using only archival histologic slides, accurately predicted postoperative recurrence in PDAC and revealed image features linked to outcomes and gene expression.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw and processed expression data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE300858.
Code availability
The Python scripts used for data analysis are available at GitHub: https://github.com/takamatsuM/PancCancer_CNN-RandomForest.
References
Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10:10–27.
Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025:10–45. https://doi.org/10.3322/caac.21871.
Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, Maitra A. Pancreatic cancer: advances and challenges. Cell. 2023;186:1729–54.
Hayashi A, Hong J, Iacobuzio-Donahue CA. The pancreatic cancer genome revisited. Nat Rev Gastroenterol Hepatol. 2021;18:469–81.
Lashen AG, Wahab N, Toss M, Miligy I, Ghanaam S, Makhlouf S, et al. Characterization of breast cancer intra-tumor heterogeneity using artificial intelligence. Cancers. 2024;16:3849.
Lee JH, Song GY, Lee J, Kang SR, Moon KM, Choi YD, et al. Prediction of immunochemotherapy response for diffuse large B-cell lymphoma using artificial intelligence digital pathology. J Pathol Clin Res. 2024;10:e12370.
Calderaro J, Ghaffari Laleh N, Zeng Q, Maille P, Favre L, Pujals A, et al. Deep learning-based phenotyping reclassifies combined hepatocellular-cholangiocarcinoma. Nat Commun. 2023;14:8290.
Pan X, AbdulJabbar K, Coelho-Lima J, Grapa AI, Zhang H, Cheung AHK, et al. The artificial intelligence-based model ANORAK improves histopathological grading of lung adenocarcinoma. Nat Cancer. 2024;5:347–63.
Wei JW, Tafe LJ, Linnik YA, Vaickus LJ, Tomita N, Hassanpour S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep. 2019;9:3358.
Fu X, Liu T, Xiong Z, Smaill BH, Stiles MK, Zhao J. Segmentation of histological images and fibrosis identification with a convolutional neural network. Comput Biol Med. 2018;98:147–58.
Liao H, Xiong T, Peng J, Xu L, Liao M, Zhang Z, et al. Classification and prognosis prediction from histopathological images of hepatocellular carcinoma by a fully automated pipeline based on machine learning. Ann Surg Oncol. 2020;27:2359–69.
Jiang B, Bao L, He S, Chen X, Jin Z, Ye Y. Deep learning applications in breast cancer histopathological imaging: diagnosis, treatment, and prognosis. Breast Cancer Res. 2024;26:137.
Wang W, Chen G, Zhang W, Zhang X, Huang M, Li C, et al. The crucial prognostic signaling pathways of pancreatic ductal adenocarcinoma were identified by single-cell and bulk RNA sequencing data. Hum Genet. 2024;143:1109–29.
Barthel S, Falcomata C, Rad R, Theis FJ, Saur D. Single-cell profiling to explore pancreatic cancer heterogeneity, plasticity and response to therapy. Nat Cancer. 2023;4:454–67.
Murakawa M, Kawahara S, Takahashi D, Kamioka Y, Yamamoto N, Kobayashi S, et al. Risk factors for early recurrence in patients with pancreatic ductal adenocarcinoma who underwent curative resection. World J Surg Oncol. 2023;21:263.
Claudio Quiros A, Coudray N, Yeaton A, Yang X, Liu B, Le H, et al. Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides. Nat Commun. 2024;15:4596.
Wang Y, Ali MA, Vallon-Christersson J, Humphreys K, Hartman J, Rantalainen M. Transcriptional intra-tumour heterogeneity predicted by deep learning in routine breast histopathology slides provides independent prognostic information. Eur J Cancer. 2023;191:112953.
Gao R, Yuan X, Ma Y, Wei T, Johnston L, Shao Y, et al. Harnessing TME depicted by histological images to improve cancer prognosis through a deep learning system. Cell Rep Med. 2024;5:101536.
Inoue Y, Takamatsu M, Masugi Y, Suzuki T, Hamada T, Abe S, et al. Blood group antigen expression in blood and tumor in relation to survival outcomes in resected pancreatic cancer, overall and by adjuvant chemotherapy regimens. Ann Surg Oncol. 2025. https://doi.org/10.1245/s10434-025-17289-7.
Ishida M, Fujii T, Kishiwada M, Shibuya K, Satoi S, Ueno M, et al. Japanese classification of pancreatic carcinoma by the Japan Pancreas Society: Eighth edition. J Hepatobiliary Pancreat Sci. 2024;31:755–68.
World Health Organization. WHO classification of tumours: digestive system tumours. 5th edn. Lyon, France: International Agency for Research on Cancer; 2019.
Amin MB, Edge SB, Greene FL, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th edn. New York: Springer; 2017.
Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 8th edn. Oxford: Wiley-Blackwell; 2017.
Suzuki T, Masugi Y, Inoue Y, Hamada T, Tanaka M, Takamatsu M, et al. KRAS variant allele frequency, but not mutation positivity, associates with survival of patients with pancreatic cancer. Cancer Sci. 2022;113:3097–109.
Masugi Y, Takamatsu M, Tanaka M, Hara K, Inoue Y, Hamada T, et al. Post-operative mortality and recurrence patterns in pancreatic cancer according to KRAS mutation and CDKN2A, p53, and SMAD4 expression. J Pathol Clin Res. 2023;9:339–53.
Hayashi K, Ono Y, Takamatsu M, Oba A, Ito H, Sato T, et al. Prediction of recurrence pattern of pancreatic cancer post-pancreatic surgery using histology-based supervised machine learning algorithms: a single-center retrospective study. Ann Surg Oncol. 2022:4624–34. https://doi.org/10.1245/s10434-022-11471-x.
Howard A, Sandler M, Chu G, Chen L-C, Chen B, Tan M, et al. MobileNetV3: searching for mobile architecture. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV). IEEE; 2019. pp. 1314–24.
Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. In: Proceedings of the 36th International Conference on Machine Learning (ICML). Proc Mach Learn Res. 2019;97:6105–14.
Breiman L, FJ, Olshen RA, Stone CJ. Classification and regression trees. Belmont, CA: Wadsworth International Group; 1984.
Browning L, Jesus C, Malacrino S, Guan Y, White K, Puddle A, et al. Artificial intelligence-based quality assessment of histopathology whole-slide images within a clinical workflow: assessment of ‘pathprofiler’ in a diagnostic pathology setting. Diagnostics. 2024;14:990.
Polónia A, Campelos S, Ribeiro A, Aymore I, Pinto D, Biskup-Fruzynska M, et al. Artificial intelligence improves the accuracy in histologic classification of breast lesions. Am J Clin Pathol. 2021;155:527–36.
Shafi S, Parwani AV. Artificial intelligence in diagnostic pathology. Diagn Pathol. 2023;18:109.
Zhang Y, Yang Z, Chen R, Zhu Y, Liu L, Dong J, et al. Histopathology images-based deep learning prediction of prognosis and therapeutic response in small cell lung cancer. NPJ Digit Med. 2024;7:15.
Yang H, Li W, Ren L, Yang Y, Zhang Y, Ge B, et al. Progress on diagnostic and prognostic markers of pancreatic cancer. Oncol Res. 2023;31:83–99.
Yang J, Zhou H, Li H, Zhao F, Tong K. Nomogram incorporating prognostic immune-inflammatory-nutritional score for survival prediction in pancreatic cancer: a retrospective study. BMC Cancer. 2024;24:193.
Liu B, Fu T, He P, Du C, Xu K. Construction of a five-gene prognostic model based on immune-related genes for the prediction of survival in pancreatic cancer. Biosci Rep. 2021;41:20204301.
Groot VP, Gemenetzis G, Blair AB, Rivero-Soto RJ, Yu J, Javed AA, et al. Defining and predicting early recurrence in 957 patients with resected pancreatic ductal adenocarcinoma. Ann Surg. 2019;269:1154–62.
Wang L, Liu Z, Liang R, Wang W, Zhu R, Li J, et al. Comprehensive machine-learning survival framework develops a consensus model in large-scale multicenter cohorts for pancreatic cancer. Elife. 2022;11:e80150.
Toriola AT, Ziegler M, Li Y, Pollak M, Stolzenberg-Solomon R. Prediagnosis circulating insulin-like growth factors and pancreatic cancer survival. Ann Surg Oncol. 2017;24:3212–9.
Huang XY, Huang ZL, Yang JH, Xu YH, Sun JS, Zheng Q, et al. Pancreatic cancer cell-derived IGFBP-3 contributes to muscle wasting. J Exp Clin Cancer Res. 2016;35:46.
Hirakawa T, Yashiro M, Murata A, Hirata K, Kimura K, Amano R, et al. IGF-1 receptor and IGF binding protein-3 might predict prognosis of patients with resectable pancreatic cancer. BMC Cancer. 2013;13:392.
Yoneyama T, Ohtsuki S, Honda K, Kobayashi M, Iwasaki M, Uchida Y, et al. Identification of IGFBP2 and IGFBP3 as compensatory biomarkers for CA19-9 in early-stage pancreatic cancer using a combination of antibody-based and LC-MS/MS-based proteomics. PLoS ONE. 2016;11:e0161009.
Karagiannidis I, Jerman SJ, Jacenik D, Phinney BB, Yao R, Prossnitz ER, et al. G-CSF and G-CSFR modulate CD4 and CD8 T cell responses to promote colon tumor growth and are potential therapeutic targets. Front Immunol. 2020;11:1885.
Huang M, Zhang L, Wu Y, Zhou X, Wang Y, Zhang J, et al. CSF3R as a potential prognostic biomarker and immunotherapy target in glioma. Cent Eur J Immunol. 2024;49:155–68.
Acknowledgements
We would like to thank the following collaborators for their valuable support in tissue processing and/or data collection: Satoko Baba, Shuhei Ishii and Motoyoshi Iwakoshi, Department of Pathology, Cancer Institute Hospital, Japanese Foundation for Cancer Research, Tokyo, Japan; Kikuko Kaji, Department of Hepato-Biliary-Pancreatic Medicine, Cancer Institute Hospital, Japanese Foundation for Cancer Research, Tokyo, Japan; Kei Sakuma, Department of Pathology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan; Noriko Koga, Hepato-Biliary-Pancreatic Surgery Division, Department of Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan; and the staff of the Fourth Laboratory of Department of Pathology in Keio University School of Medicine, Tokyo, Japan.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP21K15393 to M Tak, JP20K07414 to YM, JP19K08362 and JP22H02841 to TH, JP21K15368 to TS and JP23K15485 to TT), by the Practical Research for Innovative Cancer Control Program from AMED (23ck0106807 to YN), and by grants from The Vehicle Racing Commemorative Foundation (to. M Tak), Takeda Science Foundation (to TH), Daiwa Securities Health Foundation (to TT) and Pancreas Research Foundation of Japan (to TT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
M Tak conceived and designed the study. M Tan, YM, YI, KN, YS and KS contributed to data acquisition, data interpretation and revising the article. HK and TL performed the experiments, collected data and contributed to the interpretation of the results. Y Kaw, Y Kaz, YN, TS, KH, Y Ku and TT contributed to the acquisition of external validation data, data interpretation and manuscript revision. TH contributed to data acquisition, data interpretation, drafting and revising the article. NS, YU, SU, MF, KH and MK contributed to data interpretation, manuscript revision and supervision of the study. YT, SS, TU and KT contributed to data interpretation and supervision of the study.
Corresponding authors
Ethics declarations
Competing interests
YM and TH acknowledge research funding from the Daiichi Sankyo TaNeDS Funding Program. This work was not funded by this company. No other conflicts of interest exist. The other authors declare no conflicts of interest.
Ethics approval and consent to participate
This study was designed and conducted in accordance with the Declaration of Helsinki guidelines. Given the retrospective design of the study, informed consent was obtained from all patients on an opt-out basis. The study was approved by the ethics committee at each participating center and registered with the UMIN registry (registration number: UMIN000044027).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Takamatsu, M., Tanaka, M., Masugi, Y. et al. Prognostic model for pancreatic cancer based on machine learning of routine slides and transcriptomic tumor analysis. Br J Cancer (2026). https://doi.org/10.1038/s41416-025-03308-7
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41416-025-03308-7