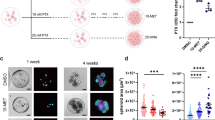
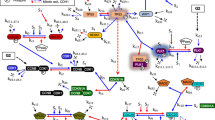
Abstract
Background
Polyploid giant cancer cells (PGCCs), characterized by enlarged or multiple nuclei, have long been considered non-proliferative and hallmarks of high malignancy. However, their functional contribution to tumor progression remains unclear.
Methods
We identified and characterized a subset of mitotically active (MA)-PGCCs in human oral squamous cell carcinoma specimens and cell lines. Mitotic activity and cell cycle was assessed using immunofluorescence, time-lapse microscopy and FUCCI. We evaluated the interactions between MA-PGCCs and cancer-associated fibroblasts (CAFs), focusing on transforming growth factor-beta (TGF-β) signaling. Chemoresistance to 5-fluorouracil (5-FU) was analyzed using cell viability assays.
Results
MA-PGCCs exhibited both bipolar and multipolar mitosis, generating heterogeneous progeny that contributed to genomic instability. These cells increased the number of CAFs with elevated TGF-β expression, promoting epithelial-mesenchymal transition (EMT) and enhancing resistance to 5-FU. Mechanistically, enhanced reactive oxygen species in MA-PGCCs upregulated urokinase-type plasminogen activator (uPA) and its receptor uPAR, promoting plasmin-mediated activation of TGF-β secreted from adjacent CAFs. Upregulation of TGF-β receptors in MA-PGCCs further amplified TGF-β signaling, accelerating EMT.
Conclusions
Our findings identify MA-PGCCs as a proliferative subpopulation that promotes EMT and chemoresistance through a TGF-β-uPA/uPAR feedback loop. Targeting this pathway may offer a novel therapeutic strategy for the treatment of aggressive tumors enriched in MA-PGCCs.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Song Y, Zhao Y, Deng Z, Zhao R, Huang Q. Stress-induced polyploid giant cancer cells: unique way of formation and non-negligible characteristics. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.724781.
Molony P, Werner R, Martin C, Callanan D, Sheahan P, Heffron C, et al. Tumour cell anaplasia and multinucleation as prognosticators in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2020. https://doi.org/10.1007/s12105-019-01081-7.
Liu HT, Xia T, You YW, Zhang QC, Ni HS, Liu YF, et al. Characteristics and clinical significance of polyploid giant cancer cells in laryngeal carcinoma. Laryngoscope Investig Otolaryngol. 2021. https://doi.org/10.1002/lio2.667.
Ogden A, Rida PC, Knudsen BS, Kucuk O, Aneja R Docetaxel-induced polyploidization may underlie chemoresistance and disease relapse. Cancer Lett. 2015. https://doi.org/10.1016/j.canlet.2015.06.025.
Matsuura T, Ueda Y, Harada Y, Hayashi K, Horisaka K, Yano Y, et al. Histological diagnosis of polyploidy discriminates an aggressive subset of hepatocellular carcinomas with poor prognosis. Br J Cancer. 2023;129:1251–60. https://doi.org/10.1038/s41416-023-02408-6.
Zhang Z, Yang J, Maimaitiyimin R, Ma M, Zhang H, Wang R. Radiation-induced mitotic catastrophe is associated with down-regulated ribosomal biosynthesis and mitosis genes. All Life. 2020;13:474–85. https://doi.org/10.1080/26895293.2020.1806117.
Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33:116–28. https://doi.org/10.1038/onc.2013.96.
Nehme Z, Pasquereau S, Haidar Ahmad S, Coaquette A, Molimard C, Monnien F, et al. Polyploid giant cancer cells, stemness and epithelial-mesenchymal plasticity elicited by human cytomegalovirus. Oncogene. 2021;40:3030–46. https://doi.org/10.1038/s41388-021-01715-7.
Puig PE, Guilly MN, Bouchot A, Droin N, Cathelin D, Bouyer F, et al. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int. 2008;32:1031–43. https://doi.org/10.1016/j.cellbi.2008.04.021.
Zhang J, Qiao Q, Xu H, Zhou R, Liu X. Human cell polyploidization: the good and the evil. Semin Cancer Biol. 2022;81:54–63. https://doi.org/10.1016/j.semcancer.2021.04.005.
Richards JS, Candelaria NR, Lanz RB. Polyploid giant cancer cells and ovarian cancer: new insights into mitotic regulators and polyploidy. Biol Reprod. 2021;105:305–16. https://doi.org/10.1093/biolre/ioab102.
Liu P, Wang L, Yu H. Polyploid giant cancer cells: origin, possible pathways of formation, characteristics, and mechanisms of regulation. Front Cell Dev Biol. 2024;12:1410637 https://doi.org/10.3389/fcell.2024.1410637.
Shabo I, Svanvik J, Lindström A, Lechertier T, Trabulo S, Hulit J, et al. Roles of cell fusion, hybridization and polyploid cell formation in cancer metastasis. World J Clin Oncol. 2020;11:121–35. https://doi.org/10.5306/wjco.v11.i3.121.
White-Gilbertson S, Voelkel-Johnson C. Giants and monsters: Unexpected characters in the story of cancer recurrence. Adv Cancer Res. 2022;148:201–232. https://doi.org/10.1016/bs.acr.2020.03.001.
Illidge TM, Cragg MS, Fringes B, Olive P, Erenpreisa JA. Polyploid giant cells provide a survival mechanism for p53 mutant cells after DNA damage. Cell Biol Int. 2000;24:621–33. https://doi.org/10.1006/cbir.2000.0557.
Zhang S, Mercado-Uribe I, Sood A, Bast RC, Liu J. Coevolution of neoplastic epithelial cells and multilineage stroma via polyploid giant cells during immortalization and transformation of mullerian epithelial cells. Genes Cancer. 2016;7:60–72. https://doi.org/10.18632/genesandcancer.102.
Zhang S, Mercado-Uribe I, Liu J. Tumor stroma and differentiated cancer cells can be originated directly from polyploid giant cancer cells induced by paclitaxel. Int J Cancer. 2014;134:508–18. https://doi.org/10.1002/ijc.28319.
Zhang D, Yang X, Yang Z, Fei F, Li S, Qu J, et al. Daughter cells and erythroid cells budding from PGCCs and their clinicopathological significances in colorectal cancer. J Cancer. 2017;8:469–78. https://doi.org/10.7150/jca.17012.
Zhang S, Mercado-Uribe I, Hanash S, Liu J. iTRAQ-based proteomic analysis of polyploid giant cancer cells and budding progeny cells reveals several distinct pathways for ovarian cancer development. PLoS ONE. 2013;8:80120 https://doi.org/10.1371/journal.pone.0080120.
Linares J, Marín-Jiménez JA, Badia-Ramentol J, Calon A. Determinants and functions of CAFs secretome during cancer progression and therapy. Front Cell Dev Biol. 2021;8. https://doi.org/10.3389/fcell.2020.621070.
Zhao Z, Li T, Yuan Y, Zhu Y. What is new in cancer-associated fibroblast biomarkers?. Cell Commun Signal. 2023;21:96. https://doi.org/10.1186/s12964-023-01125-0.
Brichkina A, Polo P, Sharma SD, Visestamkul N, Lauth M. A quick guide to CAF subtypes in pancreatic cancer. Cancers. 2023;15:2614 https://doi.org/10.3390/cancers15092614.
Pereira BA, Vennin C, Papanicolaou M, Chambers CR, Herrmann D, Morton JP, et al. CAF Subpopulations: a new reservoir of stromal targets in pancreatic cancer. Trends Cancer. 2019;5:724–41. https://doi.org/10.1016/j.trecan.2019.09.010.
Luong T, Golivi Y, Nagaraju GP, El-Rayes BF. Fibroblast heterogeneity in pancreatic ductal adenocarcinoma: perspectives in immunotherapy. Cytokine Growth Factor Rev. 2022;68:107–15. https://doi.org/10.1016/j.cytogfr.2022.09.001.
Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282–301. https://doi.org/10.1158/2159-8290.CD-18-0710.
Fang L, Che Y, Zhang C, Huang J, Lei Y, Lu Z, et al. LAMC1 upregulation via TGFβ induces inflammatory cancer-associated fibroblasts in esophageal squamous cell carcinoma via NF-κB-CXCL1-STAT3. Mol Oncol. 2021;15:3125–46. https://doi.org/10.1002/1878-0261.13053.
Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–23. https://doi.org/10.1158/2159-8290.CD-19-0094.
Itoh G, Ikeda M, Iemura K, Amin MA, Kuriyama S, Tanaka M, et al. Lateral attachment of kinetochores to microtubules is enriched in prometaphase rosette and facilitates chromosome alignment and bi-orientation establishment. Sci Rep. 2018;8:3888. https://doi.org/10.1038/s41598-018-22164-5.
Zhang Z, Aung KM, Uhlin BE, Wai SN. Reversible senescence of human colon cancer cells after blockage of mitosis/cytokinesis caused by the CNF1 cyclomodulin from Escherichia coli. Sci Rep. 2018;8:17780. https://doi.org/10.1038/s41598-018-36036-5.
Legrand AJ, Poletto M, Pankova D, Clementi E, Moore J, Castro-Giner F, et al. Persistent DNA strand breaks induce a CAF-like phenotype in normal fibroblasts. Oncotarget. 2018;9:13666–81. https://doi.org/10.18632/oncotarget.24446.
Laurenzana A, Biagioni A, Bianchini F, Peppicelli S, Chillà A, Margheri F, et al. Inhibition of uPAR-TGFβ crosstalk blocks MSC-dependent EMT in melanoma cells. J Mol Med. 2015;93:783–94. https://doi.org/10.1007/s00109-015-1266-2.
Santibanez JF, Obradović H, Kukolj T, Krstić J. Transforming growth factor-β, matrix metalloproteinases, and urokinase-type plasminogen activator interaction in the cancer epithelial to mesenchymal transition. Dev Dyn. 2018;247:382–95. https://doi.org/10.1002/dvdy.24554.
Parekh A, Das S, Parida S, Das CK, Dutta D, Mallick SK, et al. Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo. Oncogene. 2018;37:4546–61. https://doi.org/10.1038/s41388-018-0272-6.
Kuczler MD, Olseen AM, Pienta KJ, Amend SR. ROS-induced cell cycle arrest as a mechanism of resistance in polyaneuploid cancer cells (PACCs). Prog Biophys Mol Biol. 2021;165:3–7. https://doi.org/10.1016/j.pbiomolbio.2021.05.002.
Smith ER, Wang JQ, Yang DH, Xu XX. Paclitaxel resistance related to nuclear envelope structural sturdiness. Drug Resist Updat. 2022;65:100881 https://doi.org/10.1016/j.drup.2022.100881.
Wang GF, Dong Q, Bai Y, Yuan J, Xu Q, Cao C, et al. Oxidative stress induces mitotic arrest by inhibiting Aurora A-involved mitotic spindle formation. Free Radic Biol Med. 2017;103:177–87. https://doi.org/10.1016/j.freeradbiomed.2016.12.031.
Mosieniak G, Sliwinska MA, Alster O, Strzeszewska A, Sunderland P, Piechota M, et al. Polyploidy formation in doxorubicin-treated cancer cells can favor escape from senescence. Neoplasia. 2015;17:882–93. https://doi.org/10.1016/j.neo.2015.11.008.
Li P, Zhou L, Dai Z, Jin X, Liu X, Matsumoto Y, et al. High LET radiation enhances nocodazole Induced cell death in HeLa cells through mitotic catastrophe and apoptosis. J Radiat Res. 2011;52:481–9. https://doi.org/10.1269/jrr.10186.
Tsuiki H, Nitta M, Tada M, Inagaki M, Ushio Y, Saya H. Mechanism of hyperploid cell formation induced by microtubule inhibiting drug in glioma cell lines. Oncogene. 2001;20:420–9. https://doi.org/10.1038/sj.onc.1204126.
Flores-López LA, Martínez-Hernández MG, Viedma-Rodríguez R, Díaz-Flores M, Baiza-Gutman LA. High glucose and insulin enhance uPA expression, ROS formation and invasiveness in breast cancer-derived cells. Cell Oncol. 2016;39:365–78. https://doi.org/10.1007/s13402-016-0282-8.
Lee KH, Kim SW, Kim JR. Reactive oxygen species regulate urokinase plasminogen activator expression and cell invasion via mitogen-activated protein kinase pathways after treatment with hepatocyte growth factor in stomach cancer cells. J Exp Clin Cancer Res. 2009;28:73 https://doi.org/10.1186/1756-9966-28-73.
Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018;8. https://doi.org/10.3389/fonc.2018.00024.
Moirangthem A, Bondhopadhyay B, Mukherjee M, Bandyopadhyay A, Mukherjee N, Konar K, et al. Simultaneous knockdown of uPA and MMP9 can reduce breast cancer progression by increasing cell-cell adhesion and modulating EMT genes. Sci Rep. 2016;6:21903. https://doi.org/10.1038/srep21903.
Eastman BM, Jo M, Webb DL, Takimoto S, Gonias SL. A transformation in the mechanism by which the urokinase receptor signals provides a selection advantage for estrogen receptor-expressing breast cancer cells in the absence of estrogen. Cell Signal. 2012;24:1847–55. https://doi.org/10.1016/j.cellsig.2012.05.011.
Aguirre Ghiso JA. Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo. Oncogene. 2002;21:2513–24. https://doi.org/10.1038/sj.onc.1205342.
Lv T, Zhao Y, Jiang X, Yuan H, Wang H, Cui X, et al. uPAR: an essential factor for tumor development. J Cancer. 2021;12:7026–7040. https://doi.org/10.7150/jca.62281.
Zhai BT, Tian H, Sun J, Zou JB, Zhang XF, Cheng JX, et al. Urokinase-type plasminogen activator receptor (uPAR) as a therapeutic target in cancer. J Transl Med. 2022;20:135. https://doi.org/10.1186/s12967-022-03329-3.
Chen WC, Wu CC, Liu YP, Zhuo GY, Wang YK, Chen YH, et al. Elafin as a prognostic marker in esophageal squamous cell carcinoma: a pilot study using three-dimensional imaging and genomic profiling. Cancers. 2023;15. https://doi.org/10.3390/cancers15153825.
Díaz VM, Hurtado M, Thomson TM, Reventós J, Paciucci R. Specific interaction of tissue-type plasminogen activator (t-PA) with annexin II on the membrane of pancreatic cancer cells activates plasminogen and promotes invasion in vitro. Gut. 2004;53:993–1000. https://doi.org/10.1136/gut.2003.026831.
Schöpe PC, Torke S, Kobelt D, Kortüm B, Treese C, Dumbani M, et al. MACC1 revisited- an in-depth review of a master of metastasis. Biomark Res. 2024;12:146. https://doi.org/10.1186/s40364-024-00689-4.
Zheng H, Wu X, Guo L, Liu J. MyD88 signaling pathways: role in breast cancer. Front Oncol. 2024;14:1336696 https://doi.org/10.3389/fonc.2024.1336696.
Li H, Yu Z, Wang H, Wang N, Sun X, Yang S, et al. Role of ANO1 in tumors and tumor immunity. J Cancer Res Clin Oncol. 2022;148:2045–68. https://doi.org/10.1007/s00432-022-04004-2.
Sun H, Liu K, Huang J, Sun Q, Shao C, Luo J, et al. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco Targets Ther. 2019;ume 12:2829–42. https://doi.org/10.2147/OTT.S190934.
Batool A, Liu H, Liu YX, Chen SR. CD83, a novel MAPK signaling pathway interactor, determines ovarian cancer cell fate. Cancers. 2020;12:2269 https://doi.org/10.3390/cancers12082269.
Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, et al. Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a highly sensitive biomarker for gastric cancer patients. Int J Cancer. 2009;125:2383–92. https://doi.org/10.1002/ijc.24624.
Li M, Jin S, Zhang Z, Ma H, Yang X. Interleukin-6 facilitates tumor progression by inducing ferroptosis resistance in head and neck squamous cell carcinoma. Cancer Lett. 2022;527:28–40. https://doi.org/10.1016/j.canlet.2021.12.011.
Mohamed AH, Ahmed AT, Al Abdulmonem W, Bokov DO, Shafie A, Al-Hetty HRAK, et al. Interleukin-6 serves as a critical factor in various cancer progression and therapy. Med Oncol. 2024;41:182. https://doi.org/10.1007/s12032-024-02422-5.
Beernaert B, Parkes EE. cGAS-STING signalling in cancer: striking a balance with chromosomal instability. Biochem Soc Trans. 2023;51:539–55.
Tijhuis AE, Johnson SC, McClelland SE. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol Cytogenet. 2019;12:17.
Acknowledgements
We thank members of the Bioscience Education and Research Support Center (BERSC) of Akita University for technical assistance.
Funding
This work was supported by JSPS KAKENHI grants (23K24158, 25K02514 to M. Tanaka), Takeda Science Foundation grants (to M. Tanaka and G. Itoh), a Research Grant from the Princess Takamatsu Cancer Research Fund (19-25123 to M. Tanaka), Advanced Research Grant from the Dean of the Graduate School of Medicine, Akita University (G. Itoh), the Cooperative Research Project Program of Joint Usage/Research Center at the Institute of Development, Aging and Cancer, Tohoku University (G. Itoh), and the Student Assistant of Center for Physician Scientist Training in Akita University (to K. Kanetaka).
Author information
Authors and Affiliations
Contributions
Conceptualization: GI and MT conceived the study and defined the research questions. Experiments and Investigation: GI, YF, KT1, KK, HS, YK, SK, AG, and MT conducted the experiments, collected data, and performed the necessary investigations. Methodology (Material Support): YK, KI, and KT2 provided material support, including the preparation of reagents, and assisted with experimental setup. Analysis and Interpretation of Data: GI, YF, and YK performed data analysis, with KK and AG leading the tumor tissue analysis using QuPath software. Writing—Original Draft: GI wrote the first draft of the manuscript, incorporating all contributions from co-authors. Writing—Review and Editing: MT critically reviewed and edited the manuscript. * KT1: Kurara Takagane and KT2: Kozo Tanaka.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the principles of the Declaration of Helsinki principles. This study was approved by the Akita University Ethics Committee (approval number a-1-3175, Akita, Japan).
Consent for publication
We have obtained consent from the relevant parties for the publication of the research results.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Itoh, G., Fukushi, Y., Takagane, K. et al. Effects of mitotically active polyploid giant cancer cells on chemoresistance through interaction with cancer-associated fibroblasts. Br J Cancer (2025). https://doi.org/10.1038/s41416-025-03317-6
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41416-025-03317-6