Abstract
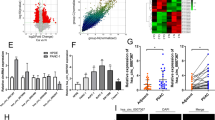
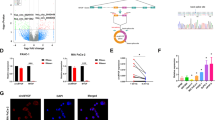
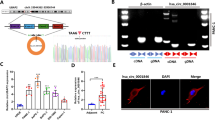
The role of circular RNAs (circRNAs) in glucose metabolism in pancreatic duct adenocarcinoma (PDAC) remains elusive. Through RNA sequencing of cells cultured under conditions of glucose deprivation, we identified hsa_circ_0007590. Sanger sequencing and RNase R and Act D treatments were performed to confirm the circular RNA features of hsa_circ_0007590. RNA in situ hybridization (RNA-ISH) and quantitative reverse transcription PCR (qRT-PCR) were used to estimate hsa_circ_0007590 expression in PDAC clinical specimens and cell lines. hsa_circ_0007590 expression was higher in PDAC patients and closely related to the clinicopathological characteristics of the disease. Cytoplasm‒nuclear fractionation and FISH assays demonstrated that hsa_circ_0007590 was located in the nucleus. Gain-of-function and loss-of-function assays were performed to assess the biological behaviors of PDAC cells. Seahorse XF assays were performed to validate the Warburg effect. hsa_circ_0007590 facilitated the proliferation, migration, and invasion of PDAC cells and promoted the Warburg effect. Mass spectrometry, RNA pulldown, RNA immunoprecipitation (RIP), RNA m6A quantification, m6A dot blot, MeRIP, and Western blotting were conducted to investigate the detailed mechanism through which hsa_circ_0007590 produces these effects. Mechanistically, hsa_circ_0007590 targeted PTBP1 and increased the expression of the m6A reader protein YTHDF2, leading to PTEN mRNA degradation and PI3K/AKT/mTOR pathway activation. Overall, hsa_circ_0007590, which targets PTBP1, reprograms glucose metabolism by attenuating the stability of m6A-modified PTEN mRNA and holds potential promise as a therapeutic target for PDAC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request. A circRNA profiling database based on RNA sequencing data from low glucose-treated and normal glucose treated MiaPaCa-2 cells was constructed in the GEO (GSE121596).
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA. 2023;73:17–48.
Wood LD, Canto MI, Jaffee EM, Simeone DM. Pancreatic cancer: pathogenesis, screening, diagnosis, and treatment. Gastroenterology. 2022;163:386–402.e1.
Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E, et al. Immunotherapy for pancreatic cancer: a 2020 update. Cancer Treatment Rev. 2020;86:102016.
Springfeld C, Ferrone CR, Katz MHG, Philip PA, Hong TS, Hackert T, et al. Neoadjuvant therapy for pancreatic cancer. Nat Rev Clin Oncol. 2023;20:318–37.
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90.
Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci USA. 2021;118:e2012881118.
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23:278–91.
Meng L, Zhang Y, Wu P, Li D, Lu Y, Shen P, et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol Cancer. 2022;21:121.
Chen Q, Li J, Shen P, Yuan H, Yin J, Ge W, et al. Biological functions, mechanisms, and clinical significance of circular RNA in pancreatic cancer: a promising rising star. Cell Biosci. 2022;12:97.
Yan B, Li X, Peng M, Zuo Y, Wang Y, Liu P, et al. The YTHDC1/GLUT3/RNF183 axis forms a positive feedback loop that modulates glucose metabolism and bladder cancer progression. Exp Mol Med. 2023;55:1145–58.
Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao S, et al. Warburg effect in colorectal cancer: the emerging roles in tumor microenvironment and therapeutic implications. J Hematol Oncol. 2022;15:160.
Liu Z, Hayashi H, Matsumura K, Ogata Y, Sato H, Shiraishi Y, et al. Hyperglycaemia induces metabolic reprogramming into a glycolytic phenotype and promotes epithelial-mesenchymal transitions via YAP/TAZ-Hedgehog signalling axis in pancreatic cancer. Br J Cancer. 2023;128:844–56.
Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178:160–75.e27.
Shaashua L, Ben-Shmuel A, Pevsner-Fischer M, Friedman G, Levi-Galibov O, Nandakumar S, et al. BRCA mutational status shapes the stromal microenvironment of pancreatic cancer linking clusterin expression in cancer associated fibroblasts with HSF1 signaling. Nat Commun. 2022;13:6513.
Liu A, Xu J. Circ_03955 promotes pancreatic cancer tumorigenesis and Warburg effect by targeting the miR-3662/HIF-1α axis. Clin Transl Oncol. 2021;23:1905–14.
Zheng D, Huang X, Peng J, Zhuang Y, Li Y, Qu J, et al. CircMYOF triggers progression and facilitates glycolysis via the VEGFA/PI3K/AKT axis by absorbing miR-4739 in pancreatic ductal adenocarcinoma. Cell Death Discov. 2021;7:362.
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695.
Shi J, Rui X, Han C, Wang C, Xu L, Jiang X. circRNF13, a novel N(6)-methyladenosine-modified circular RNA, enhances radioresistance in cervical cancer by increasing CXCL1 mRNA stability. Cell Death Discov. 2023;9:253.
Chen L, He Y, Zhu J, Zhao S, Qi S, Chen X. et al. The roles and mechanism of m(6)A RNA methylation regulators in cancer immunity. Biomed Pharmacother. 2023;163:114839
Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016–34.
Xia W, Chen W, Ni C, Meng X, Wu J, Yang Q, et al. Chemotherapy-induced exosomal circBACH1 promotes breast cancer resistance and stemness via miR-217/G3BP2 signaling pathway. Breast Cancer Res. 2023;25:85.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80.
Kumari S, Khan S, Sekhri R, Mandil H, Behrman S, Yallapu MM, et al. Protein kinase D1 regulates metabolic switch in pancreatic cancer via modulation of mTORC1. Br J Cancer. 2020;122:121–31.
Wu L, Jin Y, Zhao X, Tang K, Zhao Y, Tong L, et al. Tumor aerobic glycolysis confers immune evasion through modulating sensitivity to T cell-mediated bystander killing via TNF-α. Cell Metab. 2023;35:1580–1596.e9.
Luo H, Yu Q, Liu Y, Tang M, Liang M, Zhang D, et al. LATS kinase-mediated CTCF phosphorylation and selective loss of genomic binding. Sci Adv. 2020;6:eaaw4651.
Nguyen K, Hebert K, McConnell E, Cullen N, Cheng T, Awoyode S, et al. LKB1 signaling and patient survival outcomes in hepatocellular carcinoma. Pharmacol Res. 2023;192:106757.
Park JM, Lee DH, Kim DH. Redefining the role of AMPK in autophagy and the energy stress response. Nat Commun. 2023;14:2994.
Zhao C, Wang B, Liu E, Zhang Z. Loss of PTEN expression is associated with PI3K pathway-dependent metabolic reprogramming in hepatocellular carcinoma. Cell Commun Signal. 2020;18:131.
Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138.
Ding L, Zheng Q, Lin Y, Wang R, Wang H, Luo W, et al. Exosome-derived circTFDP2 promotes prostate cancer progression by preventing PARP1 from caspase-3-dependent cleavage. Clin Transl Med. 2023;13:e1156.
Su J, Li R, Chen Z, Liu S, Zhao H, Deng S, et al. N6-methyladenosine modification of FZR1 mRNA promotes gemcitabine resistance in pancreatic cancer. Cancer Res. 2023;83:3059–76.
Lu L, Zheng D, Qu J, Zhuang Y, Peng J, Lan S, et al. METTL16 predicts a favorable outcome and primes antitumor immunity in pancreatic ductal adenocarcinoma. Front Cell Dev Biol. 2022;10:759020.
Yao B, Zhang Q, Yang Z, An F, Nie H, Wang H, et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer. 2022;21:140.
Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–88.
Zhang Z, Wang Q, Zhao X, Shao L, Liu G, Zheng X. et al. YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 2020;11:977
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–7.
Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887–93.
Li J, Yao H, Huang J, Li C, Zhang Y, Xu R. et al. METTL3 promotes prostatic hyperplasia by regulating PTEN expression in an m(6)A-YTHDF2-dependent manner. Cell Death Dis. 2022;13:723
Acknowledgements
This study was supported by the National Natural Science Foundation of China. (No. 82203344, No. 82073150) and the Natural Science Foundation of Guangdong (No. 2021A1515010270).
Author information
Authors and Affiliations
Contributions
DZ, WC, and JP conducted the majority of the experiments and interpreted of the data. XH performed some of the experiments and collected the samples. YZ and DZ wrote the manuscript. SZ and YZ conceived the study, revised the manuscript, and were responsible for research supervision and funding acquisition. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
We declare that all research activities strictly adhere to the principles outlined in the Declaration of Helsinki. This study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (No. SYSEC-KY-KS-2019-128). After providing written informed consent, clinical samples and information were collected. The animal experiments were carried out according to the protocol approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (No. SYSU-IACUC-2021-000427).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, D., Chen, W., Peng, J. et al. Hsa_circ_0007590/PTBP1 complex reprograms glucose metabolism by reducing the stability of m6A-modified PTEN mRNA in pancreatic ductal adenocarcinoma. Cancer Gene Ther 31, 1090–1102 (2024). https://doi.org/10.1038/s41417-024-00786-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41417-024-00786-4
This article is cited by
-
Role of m6A RNA methylation regulators in pancreatic cancer: interactions and potential implications
Cancer Cell International (2025)
-
CircRNA hsa_circ_0004781 promoted cell proliferation by acting as a sponge for miR-9-5p and miR-338-3p and upregulating KLF5 and ADAM17 expression in pancreatic ductal adenocarcinoma
Cancer Cell International (2025)