Abstract
Bladder cancer (BC) is a malignancy that originates from the cells lining the bladder and is one of the most common cancers of the urinary system, capable of occurring in any part of the bladder. However, the molecular mechanisms underlying the malignant transformation of BC have not been systematically studied. This study integrated cutting-edge techniques of spatial transcriptomics (ST) and spatial metabolomics (SM) to capture the transcriptomic and metabolomic landscapes of both BC and adjacent normal tissues. ST results revealed a significant upregulation of genes associated with choline metabolism and glucose metabolism, while genes related to sphingolipid metabolism and tryptophan metabolism were significantly downregulated. Additionally, significant metabolic reprogramming was observed in BC tissues, including the upregulation of choline metabolism and glucose metabolism, as well as the downregulation of sphingolipid metabolism and tryptophan metabolism. These alterations may play a crucial role in promoting tumorigenesis and immune evasion of BC. The interpretation of ST and SM data in this study offers new insights into the molecular mechanisms underlying BC progression and provides valuable clues for the prevention and treatment of BC.

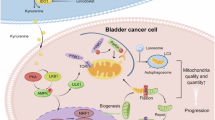
Schematic illustration of BC metabolic reprogramming.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data were presented within the article, as well as supplementary online data.
References
Compérat E, Amin MB, Cathomas R, Choudhury A, De Santis M, Kamat A, et al. Current best practice for bladder cancer: a narrative review of diagnostics and treatments. Lancet. 2022;400:1712–21.
Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: a review. JAMA. 2020;324:1980–91.
Dobruch J, Oszczudłowski M. Bladder cancer: current challenges and future directions. Medicina. 2021;57.
Woldu SL, Bagrodia A, Lotan Y. Guideline of guidelines: non-muscle-invasive bladder cancer. BJU Int. 2017;119:371–80.
Jiang DM, Gupta S, Kitchlu A, Meraz-Munoz A, North SA, Alimohamed NS, et al. Defining cisplatin eligibility in patients with muscle-invasive bladder cancer. Nat Rev Urol. 2021;18:104–14.
Lopez-Beltran A, Cookson MS, Guercio BJ, Cheng L. Advances in diagnosis and treatment of bladder cancer. BMJ. 2024;384:e076743.
Kirkali Z, Chan T, Manoharan M, Algaba F, Busch C, Cheng L, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66.
Cheung G, Sahai A, Billia M, Dasgupta P, Khan MS. Recent advances in the diagnosis and treatment of bladder cancer. BMC Med. 2013;11:13.
Tran L, Xiao J-F, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–21.
Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA: a Cancer J Clinicians. 2020;70:404–23.
Wu Z, Liu J, Dai R, Wu S. Current status and future perspectives of immunotherapy in bladder cancer treatment. Sci China Life Sci. 2021;64:512–33.
Ohshima K, Morii E. Metabolic Reprogramming of Cancer Cells during Tumor Progression and Metastasis. Metabolites. 2021;11.
Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46.
Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: Still emerging. Cell Metab. 2022;34:355–77.
Senga SS, Grose RP. Hallmarks of cancer-the new testament. Open Biol. 2021;11:200358.
Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11:5077.
Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020;9.
Robles-Remacho A, Sanchez-Martin RM, Diaz-Mochon JJ. Spatial transcriptomics: emerging technologies in tissue gene expression profiling. Anal Chem. 2023;95:15450–60.
Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–20.
Park H-E, Jo SH, Lee RH, Macks CP, Ku T, Park J, et al. Spatial transcriptomics: technical aspects of recent developments and their applications in neuroscience and cancer research. Adv Sci. 2023;10:e2206939.
Tian L, Chen F, Macosko EZ. The expanding vistas of spatial transcriptomics. Nat Biotechnol. 2023;41:773–82.
Shi Z-D, Sun Z, Zhu Z-B, Liu X, Chen J-Z, Hao L, et al. Integrated single-cell and spatial transcriptomic profiling reveals higher intratumour heterogeneity and epithelial-fibroblast interactions in recurrent bladder cancer. Clin Transl Med. 2023;13:e1338.
Gouin KH, Ing N, Plummer JT, Rosser CJ, Ben Cheikh B, Oh C, et al. An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat Commun. 2021;12:4906.
Berrell N, Sadeghirad H, Blick T, Bidgood C, Leggatt GR, O’Byrne K, et al. Metabolomics at the tumor microenvironment interface: Decoding cellular conversations. Med Res. Rev. 2024;44:1121–46.
Planque M, Igelmann S, Ferreira Campos AM, Fendt S-M. Spatial metabolomics principles and application to cancer research. Curr Opin Chem Biol. 2023;76:102362.
Ma X, Fernández FM. Advances in mass spectrometry imaging for spatial cancer metabolomics. Mass Spectrom Rev. 2024;43:235–68.
Alexandrov T, Saez-Rodriguez J, Saka SK. Enablers and challenges of spatial omics, a melting pot of technologies. Mol Syst Biol. 2023;19:e10571.
Chen C, Wang J, Pan D, Wang X, Xu Y, Yan J, et al. Applications of multi-omics analysis in human diseases. MedComm (2020). 2023;4:e315.
Du J, Yang Y-C, An Z-J, Zhang M-H, Fu X-H, Huang Z-F, et al. Advances in spatial transcriptomics and related data analysis strategies. J Transl Med. 2023;21:330.
Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell. 2022;40:920–38.
Liu X, Peng T, Xu M, Lin S, Hu B, Chu T, et al. Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications. J Hematol Oncol. 2024;17:72.
Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–37.
Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation. 2024;5:100625.
Menyhart O, Kothalawala WJ, Győrffy B. A gene set enrichment analysis for the cancer hallmarks. J Pharma Anal. 2024.
Glunde K, Penet M-F, Jiang L, Jacobs MA, Bhujwalla ZM. Choline metabolism-based molecular diagnosis of cancer: an update. Expert Rev Mol Diagnostics. 2015;15:735–47.
Li W, Li C, Liu T, Song Y, Chen P, Liu L, et al. The association of serum choline concentrations with the risk of cancers: a community-based nested case-control study. Sci Rep. 2023;13:22144.
Chen X, Qiu W, Ma X, Ren L, Feng M, Hu S, et al. Roles and mechanisms of choline metabolism in nonalcoholic fatty liver disease and cancers. Front Biosci. (Landmark Ed). 2024;29:182.
Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835–48.
De Martino S, Iorio E, Cencioni C, Aiello A, Spallotta F, Chirico M, et al. MALAT1 as a regulator of the androgen-dependent Choline Kinase A Gene in the metabolic rewiring of prostate cancer. Cancers. 2022;14.
Awwad HM, Geisel J, Obeid R. The role of choline in prostate cancer. Clin Biochem. 2012;45:1548–53.
Ceci F, Herrmann K, Hadaschik B, Castellucci P, Fanti S. Therapy assessment in prostate cancer using choline and PSMA PET/CT. Eur J Nucl Med Mol Imaging. 2017;44:78–83.
Steinberg GR, Hardie DG. New insights into activation and function of the AMPK. Nat Rev Mol Cell Biol. 2023;24:255–72.
Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–23.
Hsu C-C, Peng D, Cai Z, Lin H-K. AMPK signaling and its targeting in cancer progression and treatment. Semin Cancer Biol. 2022;85:52–68.
Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102.
Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene. 2021;40:3351–63.
Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216–37.
Kuo C-C, Wu J-Y, Wu KK. Cancer-derived extracellular succinate: a driver of cancer metastasis. J Biomed Sci. 2022;29:93.
Krzak G, Willis CM, Smith JA, Pluchino S, Peruzzotti-Jametti L. Succinate Receptor 1: An emerging regulator of myeloid cell function in inflammation. Trends Immunol. 2021;42:45–58.
Li T, Mao C, Wang X, Shi Y, Tao Y. Epigenetic crosstalk between hypoxia and tumor driven by HIF regulation. J Exp Clin Cancer Res : CR. 2020;39:224.
Wang Y-H, Yan Z-Z, Luo S-D, Hu J-J, Wu M, Zhao J, et al. Gut microbiota-derived succinate aggravates acute lung injury after intestinal ischaemia/reperfusion in mice. Eur Respir J. 2023;61.
Tufail M, Jiang C-H, Li N. Altered metabolism in cancer: insights into energy pathways and therapeutic targets. Mol Cancer. 2024;23:203.
Pajak B, Siwiak E, Sołtyka M, Priebe A, Zieliński R, Fokt I, et al. 2-Deoxy-d-glucose and its analogs: from diagnostic to therapeutic agents. Intl J Mol Sci. 2019;21.
Yang W-H, Qiu Y, Stamatatos O, Janowitz T, Lukey MJ. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer. 2021;7:790–804.
Janneh AH, Atkinson C, Tomlinson S, Ogretmen B. Sphingolipid metabolism and complement signaling in cancer progression. Trends Cancer. 2023;9:782–7.
Lee M, Lee SY, Bae Y-S. Functional roles of sphingolipids in immunity and their implication in disease. Exp Mol Med. 2023;55:1110–30.
Piazzesi A, Afsar SY, van Echten-Deckert G. Sphingolipid metabolism in the development and progression of cancer: one cancer’s help is another’s hindrance. Mol Oncol. 2021;15:3256–79.
Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18:379–401.
Xue C, Li G, Zheng Q, Gu X, Shi Q, Su Y, et al. Tryptophan metabolism in health and disease. Cell Metab. 2023;35:1304–26.
Sheridan C. IDO inhibitors move center stage in immuno-oncology. Nat Biotechnol. 2015;33:321–2.
Pham H, Torres H, Sharma P. Mental health implications in bladder cancer patients: A review. Urol Oncol. 2019;37.
Seidl C. Targets for Therapy of Bladder Cancer. Semin Nucl Med. 2020;50:162–70.
Liu R, Lee J-H, Li J, Yu R, Tan L, Xia Y, et al. Choline kinase alpha 2 acts as a protein kinase to promote lipolysis of lipid droplets. Mol Cell. 2021;81.
Li X, Hu Z, Shi Q, Qiu W, Liu Y, Liu Y, et al. Elevated choline drives KLF5-dominated transcriptional reprogramming to facilitate liver cancer progression. Oncogene. 2024;43:3121–36.
Lv W, Shi L, Pan J, Wang S. Comprehensive prognostic and immunological analysis of CCT2 in pan-cancer. Front Oncol. 2022;12:986990.
Liu W, Lu Y, Yan X, Lu Q, Sun Y, Wan X, et al. Current understanding on the role of CCT3 in cancer research. Front Oncol. 2022;12:961733.
Wettersten HI, Aboud OA, Lara PN, Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–9.
Bougeret C, Jiang S, Keydar I, Avraham H. Functional analysis of Csk and CHK Kinases in breast cancer cells*. J Biol Chem. 2001;276:33711–20.
Lacal JC, Perona R, de Castro J, Cebrián A. Choline Kinase α Inhibitors MN58b and RSM932A enhances the antitumor response to Cisplatin in lung tumor cells. Pharmaceutics. 2022;14.
Mariotto E, Bortolozzi R, Volpin I, Carta D, Serafin V, Accordi B, et al. EB-3D a novel choline kinase inhibitor induces deregulation of the AMPK-mTOR pathway and apoptosis in leukemia T-cells. Biochem Pharm. 2018;155:213–23.
de la Cueva A, Ramírez de Molina A, Alvarez-Ayerza N, Ramos MA, Cebrián A, Del Pulgar TG, et al. Combined 5-FU and ChoKα inhibitors as a new alternative therapy of colorectal cancer: evidence in human tumor-derived cell lines and mouse xenografts. PLoS One. 2013;8:e64961.
Treglia G, Piccardo A, Imperiale A, Strobel K, Kaufmann PA, Prior JO, et al. Diagnostic performance of choline PET for detection of hyperfunctioning parathyroid glands in hyperparathyroidism: a systematic review and meta-analysis. Eur J Nucl Med Mol Imaging. 2019;46:751–65.
Castellucci P, Ceci F, Fanti S. Imaging of prostate cancer using 11C-Choline PET/Computed Tomography. Urol Clin North Am. 2018;45:481–7.
Hodolič M. Imaging of prostate cancer using 18F-Choline PET/Computed Tomography. PET Clin. 2017;12:173–84.
Schmilovitz-Weiss H, Boltin D, Groshar D, Domachevsky L, Rosenbaum E, Issa N, et al. [¹¹C] choline as a potential PET/CT biomarker of liver cirrhosis: A prospective pilot study. Digest Liver Dis. 2021;53:753–9.
Alkafaas SS, Elsalahaty MI, Ismail DF, Radwan MA, Elkafas SS, Loutfy SA, et al. The emerging roles of sphingosine 1-phosphate and SphK1 in cancer resistance: a promising therapeutic target. Cancer Cell Int. 2024;24:89.
Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. 2021;20:28.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80.
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47.
Sun C, Wang A, Zhou Y, Chen P, Wang X, Huang J, et al. Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer. Nat Commun. 2023;14:2692.
Zhi Y, Wang Q, Zi M, Zhang S, Ge J, Liu K, et al. Spatial transcriptomic and metabolomic landscapes of oral submucous fibrosis-derived oral squamous cell carcinoma and its tumor microenvironment. Adv Sci. 2024;11:e2306515.
Meng W, Hao Y, He C, Li L, Zhu G. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer. 2019;18:57.
Chen Z, Han F, Du Y, Shi H, Zhou W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct Target Ther. 2023;8:70.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82473955, 82173842), the Fundamental Research Funds for the Central Universities (2632025TD04), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. We would like to thank the Wuhan Metware Biotechnology Co., Ltd. (Wuhan, China) for spatial multi-omics analysis.
Author information
Authors and Affiliations
Contributions
Lufeng Zheng designed the research. Yu Lu, Fangdie Ye, Xuedan Han and Zihan Wang analyzed the data. Yu Lu and Fangdie Ye performed the research. Yu Lu and Fangdie Ye wrote the paper. Lufeng Zheng and Xiaoman Li reviewed this paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, Y., Ye, F., Han, X. et al. Integrated spatial transcriptome and metabolism study reveals metabolic heterogeneity in human bladder cancer. Cancer Gene Ther 32, 1177–1190 (2025). https://doi.org/10.1038/s41417-025-00947-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41417-025-00947-z