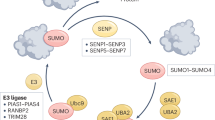
Abstract
Regulation of transcription is fundamental to the control of cellular gene expression and function. Although recent studies have revealed a role for the oncoprotein MYC in amplifying global transcription, little is known as to how the global transcription is suppressed. Here we report that SUMO and MYC mediate opposite effects upon global transcription by controlling the level of CDK9 sumoylation. On one hand, SUMO suppresses global transcription via sumoylation of CDK9, the catalytic subunit of P-TEFb kinase essential for productive transcriptional elongation. On the other hand, MYC amplifies global transcription by antagonizing CDK9 sumoylation. Sumoylation of CDK9 blocks its interaction with Cyclin T1 and thus the formation of active P-TEFb complex. Transcription profiling analyses reveal that SUMO represses global transcription, particularly of moderately to highly expressed genes and by generating a sumoylation-resistant CDK9 mutant, we confirm that sumoylation of CDK9 inhibits global transcription. Together, our data reveal that SUMO and MYC oppositely control global gene expression by regulating the dynamic sumoylation and desumoylation of CDK9.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015).
Roeder, R. G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915 (2005).
Liu, X., Bushnell, D. A. & Kornberg, R. D. RNA polymerase II transcription: structure and mechanism. Biochim. Biophys. Acta 1829, 2–8 (2013).
Hochheimer, A. & Tjian, R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17, 1309–1320 (2003).
Guo, J. & Price, D. H. RNA polymerase II transcription elongation control. Chem. Rev. 113, 8583–8603 (2013).
Jonkers, I. & Lis, J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015).
Zhou, Q., Li, T. & Price, D. H. RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 (2012).
Adelman, K. & Lis, J. T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012).
Yamaguchi, Y., Shibata, H. & Handa, H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829, 98–104 (2013).
Marshall, N. F. & Price, D. H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338 (1995).
Wada, T. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998).
Yamaguchi, Y. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999).
Cheng, B. & Price, D. H. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282, 21901–21912 (2007).
Marshall, N. F., Peng, J., Xie, Z. & Price, D. H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271, 27176–27183 (1996).
Gu, B., Eick, D. & Bensaude, O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 41, 1591–1603 (2013).
Jang, M. K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 (2005).
Nguyen, V. T., Kiss, T., Michels, A. A. & Bensaude, O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325 (2001).
Yang, Z., Zhu, Q., Luo, K. & Zhou, Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322 (2001).
Luo, Z., Lin, C. & Shilatifard, A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012).
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
Sano, M. et al. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8, 1310–1317 (2002).
Lin, C. Y. et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Rahl, P. B. et al. c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010).
Hay, R. T. Decoding the SUMO signal. Biochem. Soc. Trans. 41, 463–473 (2013).
Gareau, J. R. & Lima, C. D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 (2010).
Yang, X. J. & Chiang, C. M. Sumoylation in gene regulation, human disease, and therapeutic action. F1000Prime Rep. 5, 45 (2013).
Nayak, A. & Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15, 422 (2014).
Gill, G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541 (2005).
Ouyang, J. & Gill, G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 4, 440–444 (2009).
Yang, S. H. & Sharrocks, A. D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13, 611–617 (2004).
Rosonina, E., Duncan, S. M. & Manley, J. L. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24, 1242–1252 (2010).
Neyret-Kahn, H. et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 23, 1563–1579 (2013).
Niskanen, E. A. et al. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 16, 153 (2015).
Poukka, H., Karvonen, U., Janne, O. A. & Palvimo, J. J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA 97, 14145–14150 (2000).
Nishida, T. & Yasuda, H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 277, 41311–41317 (2002).
Wong, J., Shi, Y. B. & Wolffe, A. P. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev. 9, 2696–2711 (1995).
Huang, Z. Q., Li, J., Sachs, L. M., Cole, P. A. & Wong, J. A role for cofactor-cofactor and cofactor-histone interactions in targetingp300, SWI/SNF and mediator for transcription. EMBO J. 22, 2146–2155 (2003).
Chao, S. H. et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275, 28345–28348 (2000).
Rossi, D. J. et al. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 20, 2844–2856 (2001).
Larochelle, S. et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 19, 1108–1115 (2012).
Bartkowiak, B. et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 (2010).
Devaiah, B. N. et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl Acad. Sci. USA 109, 6927–6932 (2012).
Shore, S. M., Byers, S. A., Dent, P. & Price, D. H. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene 350, 51–58 (2005).
Cheng, J., Kang, X., Zhang, S. & Yeh, E. T. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131, 584–595 (2007).
Kang, X. et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 (2010).
Hendriks, I. A. & Vertegaal, A. C. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 (2016).
Peng, J., Marshall, N. F. & Price, D. H. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273, 13855–13860 (1998).
Bres, V., Yoh, S. M. & Jones, K. A. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20, 334–340 (2008).
Laitem, C. et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat. Struct. Mol. Biol. 22, 396–403 (2015).
Loven, J. et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Gill, G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059 (2004).
Cubenas-Potts, C. & Matunis, M. J. SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell 24, 1–12 (2013).
Chymkowitch, P., Nguea, P. A. & Enserink, J. M. SUMO-regulated transcription: challenging the dogma. BioEssays 37, 1095–1105 (2015).
Makhnevych, T. et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33, 124–135 (2009).
Tammsalu, T. et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal 7, rs2 (2014).
Diribarne, G. & Bensaude, O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 6, 122–128 (2009).
Blank, M. F. et al. SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 45, 2675–2686 (2017).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Sabo, A. et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014).
Shi, G. et al. PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1. J. Biol. Chem. 289, 29691–29700 (2014).
Fang, L. et al. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 (2014).
Haukenes, G., Szilvay, A. M., Brokstad, K. A., Kanestrom, A. & Kalland, K. H. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent (DOTAP). BioTechniques 22, 308–312 (1997).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 9, R137 (2008).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Shin, H., Liu, T., Manrai, A. K. & Liu, X. S. CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606 (2009).
Acknowledgements
We thank Dr.Jinqiu Zhou for providing yeast RNA and Dr. Xin-Hua Feng for PIAS constructs. We thank members of Wong’s laboratory for valuable discussion. This study is supported by grants from the National Natural Science Foundation of China (81272287 and 30971640 to J.L. and 81530078 to J.W.), the Ministry of Science and Technology of China (2015CB910402 and 2017YFA0504201 to J.W.), the National Institutes of Health (CA103867 to C.M.C.), Cancer Prevention Research Institute of Texas (RP110471 and RP140367 to C.M.C.), and the Welch Foundation (I-1805 to C.M.C.). F.Y. was also supported by “Outstanding doctoral dissertation cultivation plan of action” from ECNU (PY2015037).
Author information
Authors and Affiliations
Contributions
F.Y., G.S., J.L. and J.W. conceived the idea for this project. F.Y., G.S., S.C., Z.W., Y.P., Z.W., D.W. designed and conducted the experiments and interpreted the data. J.C. and T.S. performed bioinformatic analysis of RNA-seq and ChIP-seq data. Y.Z performed T cell activation experiment. N.X. and J.C provided Senp1−/− and Senp2−/− MEFs and performed related experiments. L.L. carried out mass spectromic analysis of CDK9 sumoylation. S.-Y.W. and C.-M.C. performed in vitro kinase assay. J.W. and J.L. wrote the manuscript with inputs from all the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yu, F., Shi, G., Cheng, S. et al. SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 sumoylation. Cell Res 28, 670–685 (2018). https://doi.org/10.1038/s41422-018-0023-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-018-0023-9
This article is cited by
-
CircFUNDC1 interacts with CDK9 to promote mitophagy in nucleus pulposus cells under oxidative stress and ameliorates intervertebral disc degeneration
Cell Death & Disease (2025)
-
An eRNA transcription checkpoint for diverse signal-dependent enhancer activation programs
Nature Genetics (2025)
-
Pseudouridylation of 7SK by PUS7 regulates Pol II transcription elongation
Nature Communications (2025)
-
MYC promotes global transcription in part by controlling P-TEFb complex formation via DNA-binding independent inhibition of CDK9 SUMOylation
Science China Life Sciences (2023)
-
Targeting cyclin-dependent kinase 9 in cancer therapy
Acta Pharmacologica Sinica (2022)