Abstract
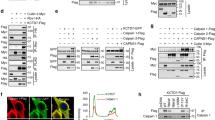
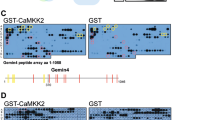
Calcium-dependent proteolytic calpains are implicated in a variety of physiological processes, as well as pathologies associated with calcium overload. However, the mechanism by which calpain is activated remains elusive since intracellular calcium levels under physiological conditions do not reach the high concentration range required to trigger calpain activation. From a candidate screening using the abundance of the calpain target glutamate receptor GluRIIA at the Drosophila neuromuscular junction as a readout, we uncovered that calpain activity was inhibited upon knockdown of Ttm50, a subunit of the Tim23 complex known to be involved in the import of proteins across the mitochondrial inner membrane. Unexpectedly, Ttm50 and calpain are co-localized at calcium stores Golgi and endoplasmic reticulum (ER), and Ttm50 interacts with calpain via its C-terminal domain. This interaction is required for calpain localization at Golgi/ER, and increases calcium sensitivity of calpain by roughly an order of magnitude. Our findings reveal the regulation of calpain activation by Ttm50, and shed new light on calpain-associated pathologies.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hanna, R. A., Campbell, R. L. & Davies, P. L. Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456, 409–412 (2008).
Strobl, S. et al. The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc. Natl. Acad. Sci. USA 97, 588–592 (2000).
Ono, Y., Saido, T. C. & Sorimachi, H. Calpain research for drug discovery: challenges and potential. Nat. Rev. Drug Discov. 15, 854–876 (2016).
Saito, K., Elce, J. S., Hamos, J. E. & Nixon, R. A. Widespread activation of calcium-activated neutral proteinase (calpain) in the brain in Alzheimer disease: a potential molecular basis for neuronal degeneration. Proc. Natl. Acad. Sci. USA 90, 2628–2632 (1993).
Lee, M. S. et al. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature 405, 360–364 (2000).
Bano, D. et al. Cleavage of the plasma membrane Na+/Ca2+ exchanger in excitotoxicity. Cell 120, 275–285 (2005).
Gan-Or, Z. et al. Mutations in CAPN1 cause autosomal-recessive hereditary spastic paraplegia. Am. J. Hum. Genet. 98, 1271 (2016).
Wang, Y. et al. Defects in the CAPN1 gene result in alterations in cerebellar development and cerebellar ataxia in mice and humans. Cell Rep. 16, 79–91 (2016).
Kanamori, T. et al. Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science 340, 1475–1478 (2013).
Metwally, E., Zhao, G., Li, W., Wang, Q. & Zhang, Y. Q. Calcium-activated calpain specifically cleaves glutamate receptor IIA but not IIB at the Drosophila neuromuscular junction. J. Neurosci. 39, 2776–2791 (2019).
Guroff, G. A neutral, calcium-activated proteinase from the soluble fraction of rat brain. J. Biol. Chem. 239, 149–155 (1964).
Berridge, M. J., Bootman, M. D. & Roderick, H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 (2003).
Clapham, D. E. Calcium signaling. Cell 131, 1047–1058 (2007).
Chan, S. L. & Mattson, M. P. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J. Neurosci. Res. 58, 167–190 (1999).
Glading, A., Lauffenburger, D. A. & Wells, A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol. 12, 46–54 (2002).
Melloni, E. et al. Acyl-CoA-binding protein is a potent m-calpain activator. J. Biol. Chem. 275, 82–86 (2000).
Lutas, A., Wahlmark, C. J., Acharjee, S. & Kawasaki, F. Genetic analysis in Drosophila reveals a role for the mitochondrial protein p32 in synaptic transmission. G3. (Bethesda) 2, 59–69 (2012).
Xiao, K., Wang, Y., Chang, Z., Lao, Y. & Chang, D. C. p32, a novel binding partner of Mcl-1, positively regulates mitochondrial Ca(2+) uptake and apoptosis. Biochem. Biophys. Res. Commun. 451, 322–328 (2014).
Koll, H. et al. Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell 68, 1163–1175 (1992).
Yamamoto, H. et al. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111, 519–528 (2002).
Cong, J., Goll, D. E., Peterson, A. M. & Kapprell, H. P. The role of autolysis in activity of the Ca2+-dependent proteinases (mu-calpain and m-calpain). J. Biol. Chem. 264, 10096–10103 (1989).
Siman, R. & Noszek, J. C. Excitatory amino acids activate calpain I and induce structural protein breakdown in vivo. Neuron 1, 279–287 (1988).
Sugiyama, S. et al. Involvement of the mitochondrial protein translocator component tim50 in growth, cell proliferation and the modulation of respiration in Drosophila. Genetics 176, 927–936 (2007).
Chen, Y. et al. An efficient two-step subcellular fractionation method for the enrichment of insulin granules from INS-1 cells. Biophys. Rep. 1, 34–40 (2015).
Lodish, H. et al. Molecular Cell Biology (4th edition, New York: W. H. Freeman press (2000).
Hood, J. L., Brooks, W. H. & Roszman, T. L. Differential compartmentalization of the calpain/calpastatin network with the endoplasmic reticulum and Golgi apparatus. J. Biol. Chem. 279, 43126–43135 (2004).
Harbauer, A. B., Zahedi, R. P., Sickmann, A., Pfanner, N. & Meisinger, C. The protein import machinery of mitochondria—a regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372 (2014).
Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell Proteom. 14, 1113–1126 (2015).
Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 (2000).
Mossmann, D., Meisinger, C. & Vogtle, F. N. Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 (2012).
Vogtle, F. N. et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009).
Shahrour, M. A. et al. Mitochondrial epileptic encephalopathy, 3-methylglutaconic aciduria and variable complex V deficiency associated with TIMM50 mutations. Clin. Genet. 91, 690–696 (2017).
Leloup, L. et al. m-Calpain activation is regulated by its membrane localization and by its binding to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 285, 33549–33566 (2010).
Meinecke, M. et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312, 1523–1526 (2006).
Collins, T. J., Berridge, M. J., Lipp, P. & Bootman, M. D. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21, 1616–1627 (2002).
Jekely, G. & Friedrich, P. Characterization of two recombinant Drosophila calpains. CALPA and a novel homolog, CALPB. J. Biol. Chem. 274, 23893–23900 (1999).
Guo, Y. et al. Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J. Biol. Chem. 279, 24813–24825 (2004).
Franco, S. J. & Huttenlocher, A. Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 (2005).
Moldoveanu, T. et al. A Ca(2+) switch aligns the active site of calpain. Cell 108, 649–660 (2002).
Sorimachi, H., Hata, S. & Ono, Y. Impact of genetic insights into calpain biology. J. Biochem. 150, 23–37 (2011).
Xu, W. et al. Calpain-mediated mGluR1alpha truncation: a key step in excitotoxicity. Neuron 53, 399–412 (2007).
Luo, L. & O’Leary, D. D. Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci. 28, 127–156 (2005).
Losonczy, A., Makara, J. K. & Magee, J. C. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature 452, 436–441 (2008).
Fontenele, M. et al. Calpain A modulates Toll responses by limited Cactus/IkappaB proteolysis. Mol. Biol. Cell 24, 2966–2980 (2013).
Bi, J. et al. Seipin promotes adipose tissue fat storage through the ER Ca(2)(+)-ATPase SERCA. Cell Metab. 19, 861–871 (2014).
Gratz, S. J. et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014).
Mao, C. X. et al. Microtubule-severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development 141, 1064–1074 (2014).
Zhao, G. et al. Drosophila S6 Kinase like inhibits neuromuscular junction growth by downregulating the BMP receptor thickveins. PLoS Genet. 11, e1004984 (2015).
Palmer, A. E. & Tsien, R. Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1, 1057–1065 (2006).
Teodoro, R. O. et al. Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst. EMBO J. 32, 2039–2055 (2013).
Wang, K. K. et al. An alpha-mercaptoacrylic acid derivative is a selective nonpeptide cell-permeable calpain inhibitor and is neuroprotective. Proc. Natl. Acad. Sci. USA 93, 6687–6692 (1996).
Li, W. et al. Angelman syndrome protein Ube3a regulates synaptic growth and endocytosis by inhibiting bmp signaling in drosophila. PLoS Genet. 12, e1006062 (2016).
Rogers, S. L. & Rogers, G. C. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3, 606–611 (2008).
Dunn, K. W., Kamocka, M. M. & McDonald, J. H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, 723–742 (2011).
Acknowledgements
This work was supported by the Strategic Priority Research Program B of the Chinese Academy of Sciences (XDBS1020100) and the National Science Foundation of China (31110103907 and 31490590) to Y.Q.Z. and (31500824) to G.Z. We thank Drs. Endre Kókai and Helena Araujo for antibodies and flies. We thank Bloomington and Tsinghua stock centers for supplying stocks. We are grateful to Dr. Yuanyuan Chen (Institute of Biophysics, CAS, China) for technical help with BLI experiment; Dr. Tao Xu and Ms. Li Min (Institute of Biophysics, CAS, China) for advice and technical assistance with fractionation experiments; Dr. Wenhua Li (Institute of Genetics and Developmental Biology (IGDB), CAS, China) for technical assistance with plasmid construction; Drs. Thomas L. Schwarz, Yuhang Chen, Xun Huang, Tieshan Tang, Yi Shi, and Dangsheng Li for discussion.
Author information
Authors and Affiliations
Contributions
E.M., G.Z., and Y.Q.Z. conceived and designed the experiments; E.M., G.Z., and Q.W. performed the experiments; E.M., G.Z., and Y.Q.Z. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Metwally, E., Zhao, G., Wang, Q. et al. Ttm50 facilitates calpain activation by anchoring it to calcium stores and increasing its sensitivity to calcium. Cell Res 31, 433–449 (2021). https://doi.org/10.1038/s41422-020-0388-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-020-0388-4
This article is cited by
-
Fam20c regulates the calpain proteolysis system through phosphorylating Calpasatatin to maintain cell homeostasis
Journal of Translational Medicine (2023)
-
Calpain activity is negatively regulated by a KCTD7–Cullin-3 complex via non-degradative ubiquitination
Cell Discovery (2023)
-
Role of CAST-Drp1 Pathway in Retinal Neuron-Regulated Necrosis in Experimental Glaucoma
Current Medical Science (2023)
-
Improved analysis method of neuromuscular junction in Drosophila larvae by transmission electron microscopy
Anatomical Science International (2022)