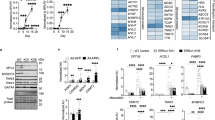
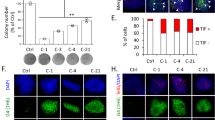
Abstract
Estrogen-related receptors (ERRα/β/γ) are orphan nuclear receptors that function in energy-demanding physiological processes, as well as in development and stem cell maintenance, but mechanisms underlying target gene activation by ERRs are largely unknown. Here, reconstituted biochemical assays that manifest ERR-dependent transcription have revealed two complementary mechanisms. On DNA templates, ERRs activate transcription with just the normal complement of general initiation factors through an interaction of the ERR DNA-binding domain with the p52 subunit of initiation factor TFIIH. On chromatin templates, activation by ERRs is dependent on AF2 domain interactions with the cell-specific coactivator PGC-1α, which in turn recruits the ubiquitous p300 and MED1/Mediator coactivators. This role of PGC-1α may also be fulfilled by other AF2-interacting coactivators like NCOA3, which is shown to recruit Mediator selectively to ERRβ and ERRγ. Importantly, combined genetic and RNA-seq analyses establish that both the TFIIH and the AF2 interaction-dependent pathways are essential for ERRβ/γ-selective gene expression and pluripotency maintenance in embryonic stem cells in which NCOA3 is a critical coactivator.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Roeder, R. G. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp. Quant. Biol. 63, 201–218 (1998).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Dancy, B. M. & Cole, P. A. Protein lysine acetylation by p300/CBP. Chem. Rev. 115, 2419–2452 (2015).
Albright, S. R. & Tjian, R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13 (2000).
Malik, S. & Roeder, R. G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010).
Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 (2018).
Roeder, R. G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 (1996).
Orphanides, G., Lagrange, T. & Reinberg, D. The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683 (1996).
Huss, J. M., Garbacz, W. G. & Xie, W. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim. Biophys. Acta 1852, 1912–1927 (2015).
Festuccia, N., Owens, N. & Navarro, P. Esrrb, an estrogen-related receptor involved in early development, pluripotency, and reprogramming. FEBS Lett. 592, 852–877 (2018).
Giguere, V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 29, 677–696 (2008).
Robyr, D., Wolffe, A. P. & Wahli, W. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol. 14, 329–347 (2000).
Xu, J., Wu, R. C. & O’Malley, B. W. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer 9, 615–630 (2009).
Chen, W. & Roeder, R. G. Mediator-dependent nuclear receptor function. Semin. Cell. Dev. Biol. 22, 749–758 (2011).
Wu, Z. et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 (1999).
Puigserver, P. et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286, 1368–1371 (1999).
Di, W. et al. PGC-1: The energetic regulator in cardiac metabolism. Curr. Issues Mol. Biol. 28, 29–46 (2018).
Villena, J. A. & Kralli, A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol. Metab. 19, 269–276 (2008).
Chen, W., Yang, Q. & Roeder, R. G. Dynamic interactions and cooperative functions of PGC-1alpha and MED1 in TRalpha-mediated activation of the brown-fat-specific UCP-1 gene. Mol. Cell 35, 755–768 (2009).
Wallberg, A. E., Yamamura, S., Malik, S., Spiegelman, B. M. & Roeder, R. G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149 (2003).
Malik, S. & Roeder, R. G. Isolation and functional characterization of the TRAP/Mediator complex. Methods Enzymol. 364, 257–284 (2003).
Kim, J., Guermah, M. & Roeder, R. G. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503 (2010).
An, W. & Roeder, R. G. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 377, 460–474 (2004).
Sladek, R., Bader, J. A. & Giguere, V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 17, 5400–5409 (1997).
Stein, R. A. et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 68, 8805–8812 (2008).
Schreiber, S. N. et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 101, 6472–6477 (2004).
Huss, J. M., Torra, I. P., Staels, B., Giguere, V. & Kelly, D. P. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24, 9079–9091 (2004).
Mootha, V. K. et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101, 6570–6575 (2004).
Huss, J. M., Kopp, R. P. & Kelly, D. P. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 277, 40265–40274 (2002).
Schreiber, S. N., Knutti, D., Brogli, K., Uhlmann, T. & Kralli, A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha). J. Biol. Chem. 278, 9013–9018 (2003).
Savkur, R. S. & Burris, T. P. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63, 207–212 (2004).
Shao, G., Heyman, R. A. & Schulman, I. G. Three amino acids specify coactivator choice by retinoid X receptors. Mol. Endocrinol. 14, 1198–1209 (2000).
Kallen, J. et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J. Biol. Chem. 279, 49330–49337 (2004).
Bugge, A., Grontved, L., Aagaard, M. M., Borup, R. & Mandrup, S. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol. Endocrinol. 23, 794–808 (2009).
Gelman, L. et al. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J. Biol. Chem. 274, 7681–7688 (1999).
Malik, S. et al. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 24, 8244–8254 (2004).
Sonoda, J. et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 21, 1909–1920 (2007).
Zhang, X. et al. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell 19, 89–100 (2005).
Cevher, M. A. et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 (2014).
Malik, S., Baek, H. J., Wu, W. & Roeder, R. G. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell. Biol. 25, 2117–2129 (2005).
Gearhart, M. D., Holmbeck, S. M., Evans, R. M., Dyson, H. J. & Wright, P. E. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J. Mol. Biol. 327, 819–832 (2003).
Huppunen, J. & Aarnisalo, P. Dimerization modulates the activity of the orphan nuclear receptor ERRgamma. Biochem. Biophys. Res. Commun. 314, 964–970 (2004).
Misawa, A. & Inoue, S. Estrogen-related receptors in breast cancer and prostate cancer. Front. Endocrinol. 6, 83 (2015).
Sun, F. et al. Promoter-enhancer communication occurs primarily within insulated neighborhoods. Mol. Cell 73, 250–263.e5 (2019).
Percharde, M. et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 26, 2286–2298 (2012).
Yamane, M., Ohtsuka, S., Matsuura, K., Nakamura, A. & Niwa, H. Overlapping functions of Kruppel-like factor family members: targeting multiple transcription factors to maintain the naive pluripotency of mouse embryonic stem cells. Development 145, dev162404 (2018).
Gaillard, S., Dwyer, M. A. & McDonnell, D. P. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol. Endocrinol. 21, 62–76 (2007).
Kamei, Y. et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA 100, 12378–12383 (2003).
Monsalve, M. et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6, 307–316 (2000).
Luo, X. et al. Posttranslational regulation of PGC-1alpha and its implication in cancer metabolism. Int. J. Cancer 145, 1475–1483 (2019).
Fernandez-Marcos, P. J. & Auwerx, J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93, 884S–890S (2011).
Ruas, J. L. et al. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331 (2012).
Chymkowitch, P., Le May, N., Charneau, P., Compe, E. & Egly, J. M. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 30, 468–479 (2011).
Traboulsi, H., Davoli, S., Catez, P., Egly, J. M. & Compe, E. Dynamic partnership between TFIIH, PGC-1alpha and SIRT1 is impaired in trichothiodystrophy. PLoS Genet. 10, e1004732 (2014).
Compe, E. et al. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 10, 1414–1422 (2007).
Compe, E. et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol. Cell. Biol. 25, 6065–6076 (2005).
Holmbeck, S. M., Dyson, H. J. & Wright, P. E. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J. Mol. Biol. 284, 533–539 (1998).
Baumann, H. et al. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry 32, 13463–13471 (1993).
Rimel, J. K. & Taatjes, D. J. The essential and multifunctional TFIIH complex. Protein Sci. 27, 1018–1037 (2018).
Kolesnikova, O., Radu, L. & Poterszman, A. TFIIH: a multi-subunit complex at the cross-roads of transcription and DNA repair. Adv. Protein Chem. Struct. Biol. 115, 21–67 (2019).
Fishburn, J., Tomko, E., Galburt, E. & Hahn, S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl. Acad. Sci. USA 112, 3961–3966 (2015).
Kim, T. K., Ebright, R. H. & Reinberg, D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000).
Fregoso, M. et al. DNA repair and transcriptional deficiencies caused by mutations in the Drosophila p52 subunit of TFIIH generate developmental defects and chromosome fragility. Mol. Cell. Biol. 27, 3640–3650 (2007).
Malik, S., Molina, H. & Xue, Z. PIC activation through functional interplay between mediator and TFIIH. J. Mol. Biol. 429, 48–63 (2017).
Osz, J. et al. Structural basis of natural promoter recognition by the retinoid X nuclear receptor. Sci. Rep. 5, 8216 (2015).
Watson, L. C. et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat. Struct. Mol. Biol. 20, 876–883 (2013).
Barry, J. B., Laganiere, J. & Giguere, V. A single nucleotide in an estrogen-related receptor alpha site can dictate mode of binding and peroxisome proliferator-activated receptor gamma coactivator 1alpha activation of target promoters. Mol. Endocrinol. 20, 302–310 (2006).
Hutchins, A. P. et al. Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells 31, 269–281 (2013).
van den Berg, D. L. et al. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol. Cell. Biol. 28, 5986–5995 (2008).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Illingworth, R. S., Botting, C. H., Grimes, G. R., Bickmore, W. A. & Eskeland, R. PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PLoS One 7, e34848 (2012).
van den Berg, D. L. et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 (2010).
Albers, M. et al. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol. Cell. Proteomics 4, 205–213 (2005).
Dufour, C. R. et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 5, 345–356 (2007).
Nakadai, T., Fukuda, A., Shimada, M., Nishimura, K. & Hisatake, K. The RNA binding complexes NF45-NF90 and NF45-NF110 associate dynamically with the c-fos gene and function as transcriptional coactivators. J. Biol. Chem. 290, 26832–26845 (2015).
Fukuda, A. et al. Reconstitution of recombinant TFIIH that can mediate activator-dependent transcription. Genes Cells 6, 707–719 (2001).
Luo, J. et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2, 1236–1247 (2007).
Yamaguchi, T. et al. Development of an all-in-one inducible lentiviral vector for gene specific analysis of reprogramming. PLoS One 7, e41007 (2012).
Fukuda, A., Nogi, Y. & Hisatake, K. The regulatory role for the ERCC3 helicase of general transcription factor TFIIH during promoter escape in transcriptional activation. Proc. Natl. Acad. Sci. USA 99, 1206–1211 (2002).
Okamoto, T. et al. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEbeta subunit. J. Biol. Chem. 273, 19866–19876 (1998).
Tang, Z. et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 154, 297–310 (2013).
Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B. & Roeder, R. G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5, 683–693 (2000).
Kobayashi, T., Kato-Itoh, M. & Nakauchi, H. Targeted organ generation using Mixl1-inducible mouse pluripotent stem cells in blastocyst complementation. Stem Cells Dev. 24, 182–189 (2015).
Niwa, H., Burdon, T., Chambers, I. & Smith, A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060 (1998).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Guermah, M., Palhan, V. B., Tackett, A. J., Chait, B. T. & Roeder, R. G. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125, 275–286 (2006).
Shimada, M., Nakadai, T., Fukuda, A. & Hisatake, K. cAMP-response element-binding protein (CREB) controls MSK1-mediated phosphorylation of histone H3 at the c-fos promoter in vitro. J. Biol. Chem. 285, 9390–9401 (2010).
Ogryzko, V. V. et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35–44 (1998).
Shimada, M. et al. Gene-specific H1 eviction through a transcriptional activator–>p300–>NAP1–>H1 pathway. Mol. Cell 74, 268–283.e5 (2019).
Poon, R. Y., Yamashita, K., Adamczewski, J. P., Hunt, T. & Shuttleworth, J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 12, 3123–3132 (1993).
Kundu, T. K. et al. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6, 551–561 (2000).
Yu, M. et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383–1386 (2015).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Cox, W. G. & Singer, V. L. A high-resolution, fluorescence-based method for localization of endogenous alkaline phosphatase activity. J. Histochem. Cytochem. 47, 1443–1456 (1999).
Acknowledgements
This work was supported by NIH grants DK071900 and CACA234575 to R.G.R. T.N. was supported by JSPS KAKENHI (19K07651). M.S. was supported by JSPS KAKENHI (19K06482), K.I. by an NCI T32 grant (CA009673) and a JSPS fellowship for research abroad, and M.A.C. by an American Cancer Society Eastern Division — New York Cancer Research Fund Postdoctoral Fellowship. We thank Drs. Vincent Giguere for anti-ERRα antibody, Takashi Onikubo for a critical review of the manuscript, Masashi Yamaji for technical advice on CFA, Koji Hisatake for TFIIH expression constructs, and Roeder laboratory members for various assistance. We also acknowledge invaluable services from The Rockefeller University Flow Cytometry Resource Center (Svetlana Mazel, Director) and the Genomics Research Center (Connie Zhao, Director).
Author information
Authors and Affiliations
Contributions
T.N. and R.G.R. planned the project. T.N. designed and conducted all experiments. M.S. provided the chromatin system. K.I. established ERRβ-KO ESCs and managed RNA-seq analysis. C.-S.C. and K.K. performed NGS data analyses. M.A.C. prepared anti-MED30 antibody and supported experiments with Mediator. R.M. supported ChIP experiments. T.N., S.M., and R.G.R. wrote the manuscript. All authors discussed the results and commented on the manuscript. R.G.R. supervised the overall work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nakadai, T., Shimada, M., Ito, K. et al. Two target gene activation pathways for orphan ERR nuclear receptors. Cell Res 33, 165–183 (2023). https://doi.org/10.1038/s41422-022-00774-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-022-00774-z
This article is cited by
-
Nuclear receptors in health and disease: signaling pathways, biological functions and pharmaceutical interventions
Signal Transduction and Targeted Therapy (2025)
-
Network pharmacology and molecular docking: combined computational approaches to explore the antihypertensive potential of Fabaceae species
Bioresources and Bioprocessing (2024)
-
LIFR regulates cholesterol-driven bidirectional hepatocyte–neutrophil cross-talk to promote liver regeneration
Nature Metabolism (2024)
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
Transcription initiation by the ERRs: no ligand but two activation pathways
Cell Research (2023)