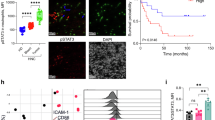
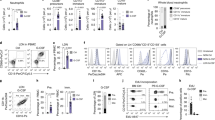
Abstract
Tumor-infiltrating neutrophils (TINs) are highly heterogeneous and mostly immunosuppressive in the tumor immune microenvironment (TIME). Current biomarkers of TINs and treatment strategies targeting TINs have not yielded optimal responses in patients across cancer types. Here, we separated human and mouse neutrophils into three developmental stages, including promyelocyte (PM), myelocyte & metamyelocyte (MC & MM), and band & segmented (BD & SC) neutrophils. Based on this separation, we observed the predominance of human but not mouse MC & MM-stage neutrophils in bone marrow (BM), which exhibit potent immunosuppressive and tumor-promoting properties. MCs & MMs also occupy the majority of TINs among patients with 17 cancer types. Moreover, through the creation of a NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt (NCG)-Gfi1−/− human immune system (HIS) mouse model, which supports efficient reconstitution of human TIN, we found a significant increase of BM MCs & MMs in tumor-bearing mice. By comparing the single-cell RNA sequencing analysis results of human neutrophils from both BM and tumors, we found that CD63 and Galectin-3 distinguish MC & MM from neutrophil populations in cancer patients. Furthermore, we proposed a strategy with Fms-like tyrosine kinase 3 ligand to specifically induce the trans-differentiation of MCs & MMs into monocytic cells, and trigger tumor control in NCG-Gfi1−/− HIS mice. Thus, our findings establish an essential role of human MC & MM-stage neutrophils in promoting cancer progression, and suggest their potential as targets for developing potential biomarkers and immunotherapies for cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
This study did not generate new materials. The scRNA-seq and bulk RNA-seq data have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences that are publicly accessible at https://ngdc.cncb.ac.cn/gsa (GSA: HRA011772, under GSA for Human category). Original code has been deposited at GitHub and is available at https://github.com/yuzu1999/Neutrophils.
References
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
Kang, H. et al. Neutrophil-macrophage communication via extracellular vesicle transfer promotes itaconate accumulation and ameliorates cytokine storm syndrome. Cell. Mol. Immunol. 21, 689–706 (2024).
Jaillon, S. et al. Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer 20, 485–503 (2020).
Shaul, M. E. & Fridlender, Z. G. Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 16, 601–620 (2019).
Hedrick, C. C. & Malanchi, I. Neutrophils in cancer: heterogeneous and multifaceted. Nat. Rev. Immunol. 22, 173–187 (2022).
Kim, R. et al. Ferroptosis of tumour neutrophils causes immune suppression in cancer. Nature 612, 338–346 (2022).
Wang, C. et al. CD300ld on neutrophils is required for tumour-driven immune suppression. Nature 621, 830–839 (2023).
Xue, R. et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612, 141–147 (2022).
Ng, M. S. F. et al. Deterministic reprogramming of neutrophils within tumors. Science 383, eadf6493 (2024).
He, X. Y. et al. Chronic stress increases metastasis via neutrophil-mediated changes to the microenvironment. Cancer Cell 42, 474–486.e12 (2024).
Benguigui, M. et al. Interferon-stimulated neutrophils as a predictor of immunotherapy response. Cancer Cell 42, 253–265.e12 (2024).
Gungabeesoon, J. et al. A neutrophil response linked to tumor control in immunotherapy. Cell 186, 1448–1464.e20 (2023).
Hirschhorn, D. et al. T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants. Cell 186, 1432–1447.e17 (2023).
Wu, Y. et al. Neutrophil profiling illuminates anti-tumor antigen-presenting potency. Cell 187, 1422–1439.e24 (2024).
Hidalgo, A., Chilvers, E. R., Summers, C. & Koenderman, L. The neutrophil life cycle. Trends Immunol. 40, 584–597 (2019).
Dinh, H. Q. et al. Coexpression of CD71 and CD117 identifies an early unipotent neutrophil progenitor population in human bone marrow. Immunity 53, 319–334 (2020).
Kwok, I. et al. Combinatorial single-cell analyses of granulocyte-monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 53, 303–318.e5 (2020).
Montaldo, E. et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat. Immunol. 23, 1470–1483 (2022).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018).
Hegde, S., Leader, A. M. & Merad, M. MDSC: markers, development, states, and unaddressed complexity. Immunity 54, 875–884 (2021).
Silvestre-Roig, C., Hidalgo, A. & Soehnlein, O. Neutrophil heterogeneity: implications for homeostasis and pathogenesis. Blood 127, 2173–2181 (2016).
Ito, R. et al. Efficient differentiation of human neutrophils with recapitulation of emergency granulopoiesis in human G-CSF knockin humanized mice. Cell Rep. 41, 111841 (2022).
Zheng, Y. et al. Human neutrophil development and functionality are enabled in a humanized mouse model. Proc. Natl. Acad. Sci. USA 119, e2121077119 (2022).
Condamine, T. et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 1, aaf8943 (2016).
Bronte, V. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016).
Veglia, F., Sanseviero, E. & Gabrilovich, D. I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 21, 485–498 (2021).
Calzetti, F. et al. CD66b−CD64dimCD115− cells in the human bone marrow represent neutrophil-committed progenitors. Nat. Immunol. 23, 679–691 (2022).
Grassi, L. et al. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018).
Liu, Z. et al. Photoactivatable Aptamer-CRISPR nanodevice enables precise profiling of interferon-gamma release in humanized mice. ACS Nano 18, 3826–3838 (2024).
Chen, X. et al. An oncolytic virus delivering tumor-irrelevant bystander T cell epitopes induces anti-tumor immunity and potentiates cancer immunotherapy. Nat. Cancer 5, 1063–1081 (2024).
Rios-Doria, J., Stevens, C., Maddage, C., Lasky, K. & Koblish, H. K. Characterization of human cancer xenografts in humanized mice. J. Immunother. Cancer 8, e000416 (2020).
Hock, H. et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003).
Karsunky, H. et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30, 295–300 (2002).
Steele, N. G. et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer 1, 1097–1112 (2020).
Guo, W. et al. Resolving the difference between left-sided and right-sided colorectal cancer by single-cell sequencing. JCI Insight 7, e152616 (2022).
Mehta, H. M. & Corey, S. J. G.-C. S. F. the guardian of granulopoiesis. Semin. Immunol. 54, 101515 (2021).
Köffel, R. et al. Monocytic cell differentiation from band-stage neutrophils under inflammatory conditions via MKK6 activation. Blood 124, 2713–2724 (2014).
Ren, D., Liu, W., Ding, S. & Li, Y. Protocol for generating human immune system mice and hydrodynamic injection to analyze human hematopoiesis in vivo. STAR Protoc. 3, 101217 (2022).
Engblom, C. et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF(high) neutrophils. Science 358, eaal5081 (2017).
Veglia, F. et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 569, 73–78 (2019).
Maas, R. R. et al. The local microenvironment drives activation of neutrophils in human brain tumors. Cell 186, 4546–4566.e27 (2023).
Zhao, Y. et al. Neutrophils resist ferroptosis and promote breast cancer metastasis through aconitate decarboxylase 1. Cell Metab. 35, 1688–1703.e10 (2023).
Bianchi, A. et al. Cell-autonomous Cxcl1 sustains tolerogenic circuitries and stromal inflammation via neutrophil-derived TNF in pancreatic cancer. Cancer Discov. 13, 1428–1453 (2023).
Monteran, L. et al. Combining TIGIT blockage with MDSC inhibition hinders breast cancer bone metastasis by activating anti-tumor immunity. Cancer Discov. 14, 1252–1275 (2024).
Wang, T. T. et al. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut 66, 1900–1911 (2017).
Gong, Z. et al. Immunosuppressive reprogramming of neutrophils by lung mesenchymal cells promotes breast cancer metastasis. Sci. Immunol. 8, eadd5204 (2023).
Barry, S. T., Gabrilovich, D. I., Sansom, O. J., Campbell, A. D. & Morton, J. P. Therapeutic targeting of tumour myeloid cells. Nat. Rev. Cancer 23, 216–237 (2023).
Aarts, C. E. M. et al. Activated neutrophils exert myeloid-derived suppressor cell activity damaging T cells beyond repair. Blood Adv. 3, 3562–3574 (2019).
Perez, C. et al. Immunogenomic identification and characterization of granulocytic myeloid-derived suppressor cells in multiple myeloma. Blood 136, 199–209 (2020).
Skokowa, J., Dale, D. C., Touw, I. P., Zeidler, C. & Welte, K. Severe congenital neutropenias. Nat. Rev. Dis. Prim. 3, 17032 (2017).
Fan, Y. et al. Differential proteomics argues against a general role for CD9, CD81 or CD63 in the sorting of proteins into extracellular vesicles. J. Extracell. Vesicles 12, e12352 (2023).
Sun, J. et al. CD63(+) cancer-associated fibroblasts confer CDK4/6 inhibitor resistance to breast cancer cells by exosomal miR-20. Cancer Lett. 588, 216747 (2024).
Tominaga, N. et al. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol. Cancer 13, 134 (2014).
Miki, Y. et al. Clinico-pathological significance of exosome marker CD63 expression on cancer cells and stromal cells in gastric cancer. PloS One 13, e0202956 (2018).
Fabre, T. et al. Identification of a broadly fibrogenic macrophage subset induced by type 3 inflammation. Sci. Immunol. 8, eadd8945 (2023).
Wang, H. et al. LGALS3 promotes treatment resistance in glioblastoma and is associated with tumor risk and prognosis. Cancer Epidemiol. Biomark. Prev. 28, 760–769 (2019).
Fan, Y. et al. Galectin-3 cooperates with CD47 to suppress phagocytosis and T-cell immunity in gastric cancer peritoneal metastases. Cancer Res. 83, 3726–3738 (2023).
Mabbitt, J. et al. Resistance to anti-PD-1/anti-PD-L1: galectin-3 inhibition with GB1211 reverses galectin-3-induced blockade of pembrolizumab and atezolizumab binding to PD-1/PD-L1. Front. Immunol. 14, 1250559 (2023).
Ruvolo, P. P. et al. LGALS3 is connected to CD74 in a previously unknown protein network that is associated with poor survival in patients with AML. EBioMedicine 44, 126–137 (2019).
Hsu, B. E. et al. Immature low-density neutrophils exhibit metabolic flexibility that facilitates breast cancer liver metastasis. Cell Rep. 27, 3902–3915.e6 (2019).
Sagiv, J. Y. et al. Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Rep. 10, 562–573 (2015).
Rice, C. M. et al. Tumour-elicited neutrophils engage mitochondrial metabolism to circumvent nutrient limitations and maintain immune suppression. Nat. Commun. 9, 5099 (2018).
Mackey, J. B. G., Coffelt, S. B. & Carlin, L. M. Neutrophil maturity in cancer. Front. Immunol. 10, 1912 (2019).
Goswami, S., Anandhan, S., Raychaudhuri, D. & Sharma, P. Myeloid cell-targeted therapies for solid tumours. Nat. Rev. Immunol. 23, 106–120 (2023).
Linde, I. L. et al. Neutrophil-activating therapy for the treatment of cancer. Cancer cell 41, 356–372.e10 (2023).
Mastio, J. et al. Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J. Exp. Med. 216, 2150–2169 (2019).
Ballesteros, I. et al. Co-option of neutrophil fates by tissue environments. Cell 183, 1282–1297.e18 (2020).
Lin, X. et al. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer 23, 108 (2024).
Zhang, H. et al. Roles of tumor-associated macrophages in anti-PD-1/PD-L1 immunotherapy for solid cancers. Mol. Cancer 22, 58 (2023).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. J. B. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. J. N. b. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. J. B. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. J. G. b. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Bardou, P., Mariette, J., Escudié, F., Djemiel, C. & Klopp, C. J. B. b. jvenn: an interactive Venn diagram viewer. BMC Bioinforma. 15, 293 (2014).
Wu, J. et al. A single-cell survey of cellular hierarchy in acute myeloid leukemia. J. Hematol. Oncol. 13, 128 (2020).
Qi, J. et al. Multimodal single-cell characterization of the human granulocyte lineage. bioRxiv https://www.biorxiv.org/content/10.1101/2021.06.12.448210v1 (2021).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018).
Velten, L. et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017).
Li, Q. scTour: a deep learning architecture for robust inference and accurate prediction of cellular dynamics. Genome Biol. 24, 149 (2023).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Acknowledgements
This work was funded by grants from the National Science and Technology Major Program (2023ZD0500400), the Fundamental Research Funds for the Central Universities (2024300408, XJ2024003602), the National Natural Science Foundation of China (32471000, U24A20378, 82373263, 82403835), the National Key R&D Program of China (2023YFC2506400), the Jiangsu Provincial Science and Technology Plan Special Fund (BK20232018) and the Funding of the Major Program of Shenzhen Bay Laboratory.
Author information
Authors and Affiliations
Contributions
Y. Li and W.L. designed the study. Y. Li, J.W., T.M. and Y. Liang supervised the study and revised the manuscript. W.L. performed the experiments and analyzed the data. with the assistance from T.S., C.L., K.C., Z.Z., Y. Luo, D.H., H.W., Shaorui L., Y.W., Shuang L., H.S., J.L., Y. Liu, D.S., S.D., H.X., L.L., J.X. and Jun X. T.S. and W.L. drafted the manuscript. The clinical samples were provided by S.D., Y. Luo and Y. Liu. The bioinformatic analyses were conducted by C.L. NCG-Gfi1−/− mice were generated by Y. Li, W.L., L.L., J.X., Jun X. and Y. Liang. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y. Li is currently consulting for GemPharmatech Co. J.W. has received research funding from Leap Therapeutics. T.M. is a consultant for Immunos Therapeutics, Daiichi Sankyo Co, TigaTx, Normunity and Pfizer. T.M. is a cofounder and equity holder of IMVAQ Therapeutics. T.M. has received research funding from Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmaceuticals, Adaptive Biotechnologies, Leap Therapeutics, and Aprea Therapeutics, and currently receives research funding from Bristol-Myers Squibb, Enterome SA, and Realta Life Sciences. T.M. is an inventor on patent applications related to work on oncolytic viral therapy, alpha virus-based vaccine, neo-antigen modeling, CD40, GITR, OX40, PD-1, and CTLA-4. The rest of authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, W., Shi, T., Lu, C. et al. Human myelocyte and metamyelocyte-stage neutrophils suppress tumor immunity and promote cancer progression. Cell Res 35, 588–606 (2025). https://doi.org/10.1038/s41422-025-01145-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-025-01145-0
This article is cited by
-
Neutrophil maturation holds the secret to human tumor suppression
Cell Research (2025)