Abstract
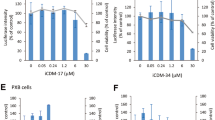
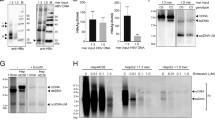
HBV infection initiates hepatitis B and promotes liver cirrhosis and hepatocellular carcinoma. IFN-α is commonly used in hepatitis B therapy, but how it inhibits HBV is not fully understood. We screened 285 human interferon-stimulated genes (ISGs) for anti-HBV activity using a cell-based assay, which revealed several anti-HBV ISGs. Among these ISGs, SAMD4A was the strongest suppressor of HBV replication. We found the binding site of SAMD4A in HBV RNA, which was a previously unidentified Smaug recognition region (SRE) sequence conserved in HBV variants. SAMD4A binds to the SRE site in viral RNA to trigger its degradation. The SAM domain in SAMD4A is critical for RNA binding and the C-terminal domain of SAMD4A is required for SAMD4A anti-HBV function. Human SAMD4B is a homolog of human SAMD4A but is not an ISG, and the murine genome encodes SAMD4. All these SAMD4 proteins suppressed HBV replication when overexpressed in vitro and in vivo. We also showed that knocking out the Samd4 gene in hepatocytes led to a higher level of HBV replication in mice and AAV-delivered SAMD4A expression reduced the virus titer in HBV-producing transgenic mice. In addition, a database analysis revealed a negative correlation between the levels of SAMD4A/B and HBV in patients. Our data suggest that SAMD4A is an important anti-HBV ISG for use in IFN therapy of hepatitis B and that the levels of SAMD4A/B expression are related to HBV sensitivity in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Perz, J. F., Armstrong, G. L., Farrington, L. A., Hutin, Y. J. F. & Bell, B. P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45, 529–538 (2006).
Yan, H. et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e00049 (2012).
Li, H. et al. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol. Immunol. 11, 175–183 (2014).
Tuttleman, J. S., Pourcel, C. & Summers, J. Formation of the pool of covalently closed circular viral-DNA in hepadnavirus-infected cells. Cell 47, 451–460 (1986).
Tong, S. P. & Revill, P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 64, S4–S16 (2016).
Newbold, J. E. et al. The covalently closed duplex form of the hepadnavirus genome exists in-situ as a heterogeneous population of viral minichromosomes. J. Virol. 69, 3350–3357 (1995).
Ghany, M. & Liang, T. J. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology 132, 1574–1585 (2007).
Perrillo, R. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49, S103–S111 (2009).
Wieland, S. F., Asabe, S., Engle, R. E., Purcell, R. H. & Chisari, F. V. Limited hepatitis B virus replication space in the chronically hepatitis C virus-infected liver. J. Virol. 88, 5184–5188 (2014).
Tan, G. Y., Song, H. X., Xu, F. C. & Cheng, G. H. When hepatitis B virus meets interferons. Front. Microbiol. 9, 1611 (2018).
Smibert, C. A., Wilson, J. E., Kerr, K. & Macdonald, P. M. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Gene Dev. 10, 2600–2609 (1996).
Aviv, T., Lin, Z., Ben-Ari, G., Smibert, C. A. & Sicheri, F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 13, 168–176 (2006).
Baez, M. V. & Boccaccio, G. L. Mammalian smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J. Biol. Chem. 280, 43131–43140 (2005).
Baez, M. V. et al. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J. Cell Biol. 195, 1141–1157 (2011).
Fernandez-Alvarez, A. J., Pascual, M. L., Boccaccio, G. L. & Thomas, M. G. Smaug variants in neural and non-neuronal cells. Commun. Integr. Biol. 9, e1139252 (2016).
de Haro, M. et al. Smaug/SAMD4A restores translational activity of CUGBP1 and suppresses CUG-induced myopathy. PLoS Genet. 9, e1003445 (2013).
Chen, Z. et al. Mutation of mouse Samd4 causes leanness, myopathy, uncoupled mitochondrial respiration, and dysregulated mTORC1 signaling. Proc. Natl Acad. Sci. USA 111, 7367–7372 (2014).
Niu, N. N. et al. RNA-binding protein SAMD4 regulates skeleton development through translational inhibition of Mig6 expression. Cell Discov. 3, 16050 (2017).
Zhang, Y. L. et al. Robust in vitro assay for analyzing the neutralization activity of serum specimens against hepatitis B virus. Emerg. Microbes Infect. 8, 724–733 (2019).
Gripon, P. et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl Acad. Sci. USA 99, 15655–15660 (2002).
Wu, Y. et al. Sleeping beauty transposon-based system for rapid generation of HBV-replicating stable cell lines. J. Virol. Methods 234, 96–100 (2016).
Zhang, T. Y. et al. Prolonged suppression of HBV in mice by a novel antibody that targets a unique epitope on hepatitis B surface antigen. Gut 65, 658–671 (2016).
Zhang, D. W. et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009).
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–U545 (2011).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–U514 (2007).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, aag2045 (2016).
Chen, Q. Y. et al. DNA-dependent activator of interferon-regulatory factors inhibits hepatitis B virus replication. World J. Gastroenterol. 18, 2850–2858 (2012).
Wang, W. H., Studach, L. L. & Andrisani, O. M. Proteins ZNF198 and SUZ12 are down-regulated in hepatitis B virus (HBV) X protein-mediated hepatocyte transformation and in HBV replication. Hepatology 53, 1137–1147 (2011).
Tavalai, N., Papior, P., Rechter, S. & Stamminger, T. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82, 126–137 (2008).
Regad, T. & Chelbi-Alix, M. K. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20, 7274–7286 (2001).
Mao, R. C. et al. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 9, e1003494 (2013).
Mao, R. C. et al. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J. Virol. 85, 1048–1057 (2011).
Liu, Y. J. et al. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 13, e1006296 (2017).
Li, J. H. et al. Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of Pre-S/S RNAs. J. Virol. 84, 6387–6399 (2010).
Wang, H. F., Kim, S. & Ryu, W. S. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 83, 5815–5824 (2009).
Nelson, M. R., Leidal, A. M. & Smibert, C. A. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 23, 150–159 (2004).
Brodsky, L. I. et al. A novel unsupervised method to identify genes important in the anti-viral response: application to interferon/ribavirin in hepatitis C patients. PLoS ONE 2, e584 (2007).
Aviv, T. et al. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 10, 614–621 (2003).
Nishimura, T. et al. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell Rep. 11, 1425–1436 (2015).
Semotok, J. L. et al. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early drosophila embryo. Curr. Biol. 15, 284–294 (2005).
Zaessinger, S., Busseau, I. & Simonelig, M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133, 4573–4583 (2006).
Huang, L. R., Wu, H. L., Chen, P. J. & Chen, D. S. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc. Natl Acad. Sci. USA 103, 17862–17867 (2006).
Yang, P. L., Althage, A., Chung, J. & Chisari, F. V. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl Acad. Sci. USA 99, 13825–13830 (2002).
Zhou, W. et al. Predictive model for inflammation grades of chronic hepatitis B: large-scale analysis of clinical parameters and gene expressions. Liver Int. 37, 1632–1641 (2017).
Hubel, P. et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 20, 493–49 (2019).
Pollack, J. R. & Ganem, D. An RNA stem-loop structure directs hepatitis-B virus genomic RNA encapsidation. J. Virol. 67, 3254–3263 (1993).
Sato, S. et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42, 123–132 (2015).
Acknowledgements
This work was supported by the National Scientific and Technological Major Project (2017ZX10202203-003), the National Natural Science Foundation of China (81788101, 31420103910, and 81630042), the 111 Project (B12001), and the National Science Foundation of China for Fostering Talents in Basic Research (J1310027).
Author information
Authors and Affiliations
Contributions
Y.W., X.F., Q.Y., N.X., and J.H. conceived and designed the experiments. Y.W., X.F., Y.S., R.L., Y.L., J.W., and X.L. performed the experiments. Y.W., X.F., Y.S., G.F., N.X., and J.H. analyzed the data. Y.W., X.F., and J.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Y., Fan, X., Song, Y. et al. SAMD4 family members suppress human hepatitis B virus by directly binding to the Smaug recognition region of viral RNA. Cell Mol Immunol 18, 1032–1044 (2021). https://doi.org/10.1038/s41423-020-0431-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41423-020-0431-x
Keywords
This article is cited by
-
LSM12 promotes the lung squamous cell carcinoma progression through mediating alternative splicing of ARRB1
Communications Biology (2025)
-
Interferon-γ-induced GBP1 is an inhibitor of human papillomavirus 18
BMC Women's Health (2024)
-
RNA binding protein SAMD4: current knowledge and future perspectives
Cell & Bioscience (2023)
-
Restoration of HBV-specific CD8+ T-cell responses by sequential low-dose IL-2 treatment in non-responder patients after IFN-α therapy
Signal Transduction and Targeted Therapy (2021)