Abstract
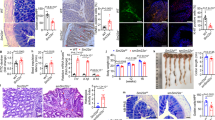
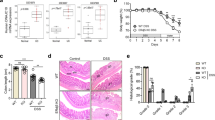
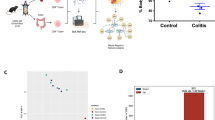
YdjC chitooligosaccharide deacetylase homolog (YDJC) has been identified as a susceptibility gene for inflammatory bowel disease (IBD), yet its role in the pathogenesis of IBD, particularly in regulating immune responses in the gut mucosa, remains elusive. In this study, we demonstrated that YDJC expression is downregulated in inflamed mucosa, particularly in the CD4+ T cells of IBD patients, and that Ydjc deficiency promotes CD4+ T-cell proliferation and Th1 cell differentiation, thereby exacerbating acute and chronic colitis in mice. Integrative transcriptomic, proteomic, and metabolomic analyses revealed that Ydjc-/-CD4+ T cells exhibit upregulated SREBP2-mediated cholesterol biosynthesis. Consistently, treatment with key enzyme inhibitors targeting cholesterol biosynthesis, including simvastatin, fatostatin, and AAV-sh-Srebf2, markedly suppressed CD4+ T-cell proliferation and Th1 cell differentiation, thereby alleviating colitis in Ydjc-/- mice. Mechanistically, YDJC directly deacetylates SREBP2, which further suppresses downstream target gene expression (e.g., Hmgcr, Hmgcs1, and Cyp51). Therefore, our findings elucidate a novel mechanism whereby YDJC restrains intestinal mucosal inflammation by downregulating SREBP2-driven Th1 cell differentiation, suggesting that targeting YDJC and SREBP2-mediated cholesterol biosynthesis may serve as promising therapeutic strategies for IBD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17. https://doi.org/10.1038/nrgastro.2015.34.
de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–61. https://doi.org/10.1038/ng.3760.
Imagawa T, Iino H, Kanagawa M, Ebihara A, Kuramitsu S, Tsuge H. Crystal structure of the YdjC-family protein TTHB029 from Thermus thermophilus HB8: structural relationship with peptidoglycan N-acetylglucosamine deacetylase. Biochem Biophys Res Commun. 2008;367:535–41. https://doi.org/10.1016/j.bbrc.2007.12.144.
Verma SC, Mahadevan S. The chbG gene of the chitobiose (chb) operon of Escherichia coli encodes a chitooligosaccharide deacetylase. J Bacteriol. 2012;194:4959–71. https://doi.org/10.1128/JB.00533-12.
Kasher M, Cherny SS, Group CIW, Livshits G. Exploring potential shared genetic influences between rheumatoid arthritis and blood lipid levels. Atherosclerosis. 2022;363:48–56. https://doi.org/10.1016/j.atherosclerosis.2022.11.006.
Ellinghaus D, Ellinghaus E, Nair RP, Stuart PE, Esko T, Metspalu A, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–47. https://doi.org/10.1016/j.ajhg.2012.02.020.
Ye BD, Choi H, Hong M, Yun WJ, Low HQ, Haritunians T, et al. Identification of ten additional susceptibility Loci for Ulcerative Colitis Through Immunochip Analysis in Koreans. Inflamm Bowel Dis. 2016;22:13–19. https://doi.org/10.1097/MIB.0000000000000584.
Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. https://doi.org/10.1038/ng.764.
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. https://doi.org/10.1038/ng.717.
Liu Z, Liu R, Gao H, Jung S, Gao X, Sun R, et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat Genet. 2023;55:796–806. https://doi.org/10.1038/s41588-023-01384-0.
Sands BE, Irving PM, Hoops T, Izanec JL, Gao LL, Gasink C, et al. Ustekinumab versus adalimumab for induction and maintenance therapy in biologic-naive patients with moderately to severely active Crohn’s disease: a multicenter, randomized, double-blind, parallel-group, phase 3b trial. Lancet. 2022;399:2200–11. https://doi.org/10.1016/S0140-6736(22)00688-2.
Chapman NM, Chi H. Metabolic adaptation of lymphocytes in immunity and disease. Immunity. 2022;55:14–30. https://doi.org/10.1016/j.immuni.2021.12.012.
Geltink RIK, Kyle RL, Pearce EL. Unraveling the complex interplay between T-cell metabolism and function. Annu Rev Immunol. 2018;36:461–88. https://doi.org/10.1146/annurev-immunol-042617-053019.
Reina-Campos M, Heeg M, Kennewick K, Mathews IT, Galletti G, Luna V, et al. Metabolic programs of T-cell tissue residency empower tumor immunity. Nature. 2023;621:179–87. https://doi.org/10.1038/s41586-023-06483-w.
Liu ZJ, Yadav PK, Su JL, Wang JS, Fei K. Potential role of Th17 cells in the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2009;15:5784–8. https://doi.org/10.3748/wjg.15.5784.
Neurath MF. Strategies for targeting cytokines in inflammatory bowel disease. Nat Rev Immunol. 2024;24:559–76. https://doi.org/10.1038/s41577-024-01008-6.
Santos AL, Preta G. Lipids in the cell: organization regulates function. Cell Mol Life Sci. 2018;75:1909–27. https://doi.org/10.1007/s00018-018-2765-4.
Ikeda M, Takeshima F, Isomoto H, Shikuwa S, Mizuta Y, Ozono Y, et al. Simvastatin attenuates trinitrobenzene sulfonic acid-induced colitis, but not oxazalone-induced colitis. Dig Dis Sci. 2008;53:1869–75. https://doi.org/10.1007/s10620-007-0102-0.
Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. https://doi.org/10.1172/JCI15593.
Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–45. https://doi.org/10.1038/s41580-019-0190-7.
Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40:439–52. https://doi.org/10.1016/s0163-7827(01)00010-8.
Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–48. https://doi.org/10.1016/j.biochi.2004.09.018.
Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–17. https://doi.org/10.1101/gad.1901210.
Gao H, Liu Z. The latest breakthrough on genetic characteristics of inflammatory bowel disease in Chinese and other East Asian ancestries. Precis Clin Med. 2023;6:pbad017. https://doi.org/10.1093/pcmedi/pbad017.
Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–43. https://doi.org/10.1016/j.immuni.2013.04.005.
Shale M, Schiering C, Powrie F. CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–82. https://doi.org/10.1111/imr.12039.
Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, et al. Spatial map of human T-cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–28. https://doi.org/10.1016/j.cell.2014.10.026.
Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T-cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–61. https://doi.org/10.1146/annurev-immunol-032712-095954.
Zhang H, Kong X, Wang W, Zhou H, Qu H, Guan Z, et al. TRIM25-mediated INSIG1 Ubiquitination Promotes MASH progression through reprogramming lipid metabolism. Adv Sci. 2025;12:2414646. https://doi.org/10.1002/advs.202414646.
Wang Y, Choe J, Shi Y, Thi TT, Cao X, Hu Y, et al. Depletion of Hepatic SREBP2 Protects against Hypercholesterolemia and Atherosclerosis through the ANGPTL3-LPL Axis.Adv Sci. 2025;12:2412677. https://doi.org/10.1002/advs.202412677.
Song X, He X, Li X, Qian Y. The roles and functional mechanisms of interleukin-17 family cytokines in mucosal immunity. Cell Mol Immunol. 2016;13:418–31. https://doi.org/10.1038/cmi.2015.105.
Ohara D, Takeuchi Y, Hirota K. Type 17 immunity: novel insights into intestinal homeostasis and autoimmune pathogenesis driven by gut-primed T cells. Cell Mol Immunol. 2024;21:1183–1200. https://doi.org/10.1038/s41423-024-01218-x.
Waddington KE, Robinson GA, Rubio-Cuesta B, Chrifi-Alaoui E, Andreone S, Poon KS, et al. LXR directly regulates glycosphingolipid synthesis and affects human CD4+ T-cell function. Proc Natl Acad Sci USA 2021:118. https://doi.org/10.1073/pnas.2017394118.
Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–99. https://doi.org/10.1038/ni.2570.
Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. https://doi.org/10.1016/j.cell.2008.04.052.
Surls J, Nazarov-Stoica C, Kehl M, Olsen C, Casares S, Brumeanu TD. Increased membrane cholesterol in lymphocytes diverts T cells toward an inflammatory response. PLoS One. 2012;7:e38733 https://doi.org/10.1371/journal.pone.0038733.
Jakobsson T, Vedin LL, Hassan T, Venteclef N, Greco D, D'Amato M, et al. The oxysterol receptor LXRbeta protects against DSS- and TNBS-induced colitis in mice. Mucosal Immunol. 2014;7:1416–28. https://doi.org/10.1038/mi.2014.31.
Greenwood J, Steinman L, Zamvil SS. Statin therapy and autoimmune disease: from protein prenylation to immunomodulation. Nat Rev Immunol. 2006;6:358–70. https://doi.org/10.1038/nri1839.
Jameel A, Ooi KG, Jeffs NR, Galatowicz G, Lightman SL, Calder VL. Statin modulation of human T-cell proliferation, IL-1beta and IL-17 production, and IFN-gamma T-cell expression: synergy with conventional immunosuppressive agents. Int J Inflam. 2013;2013:434586. https://doi.org/10.1155/2013/434586.
Ganesan A, Crum-Cianflone N, Higgins J, Qin J, Rehm C, Metcalf J, et al. High dose atorvastatin decreases cellular markers of immune activation without affecting HIV-1 RNA levels: results of a double-blind randomized placebo controlled clinical trial. J Infect Dis. 2011;203:756–64. https://doi.org/10.1093/infdis/jiq115.
Kanagarajan N, Nam JH, Noah ZA, Murthy S. Disease modifying effect of statins in dextran sulfate sodium model of mouse colitis. Inflamm Res. 2008;57:34–38. https://doi.org/10.1007/s00011-007-6177-4.
Lee JY, Kim JS, Kim JM, Kim N, Jung HC, Song IS. Simvastatin inhibits NF-kappaB signaling in intestinal epithelial cells and ameliorates acute murine colitis. Int Immunopharmacol. 2007;7:241–8. https://doi.org/10.1016/j.intimp.2006.10.013.
Aktas O, Waiczies S, Smorodchenko A, Dorr J, Seeger B, Prozorovski T, et al. Treatment of relapsing paralysis in experimental encephalomyelitis by targeting Th1 cells through atorvastatin. J Exp Med. 2003;197:725–33. https://doi.org/10.1084/jem.20021425.
Leung BP, Sattar N, Crilly A, Prach M, McCarey DW, Payne H, et al. A novel anti-inflammatory role for simvastatin in inflammatory arthritis. J Immunol. 2003;170:1524–30. https://doi.org/10.4049/jimmunol.170.3.1524.
Bhagavathula AS, Clark C, Rahmani J. Statin use and new-onset of inflammatory bowel disease: A systematic review and meta-analysis of over ten million participants. Eur J Pharm. 2021;891:173750. https://doi.org/10.1016/j.ejphar.2020.173750.
Ananthakrishnan AN, Cagan A, Cai T, Gainer VS, Shaw SY, Churchill S, et al. Statin use is associated with reduced risk of colorectal cancer in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2016;14:973–9. https://doi.org/10.1016/j.cgh.2016.02.017.
Crockett SD, Hansen RA, Stürmer T, Schectman R, Darter J, Sandler RS, et al. Statins are associated with reduced use of steroids in inflammatory bowel disease: a retrospective cohort study. Inflamm Bowel Dis. 2012;18:1048–56. https://doi.org/10.1002/ibd.21822.
Rea WE, Durrant DC, Boldy DA. Ulcerative colitis after statin treatment. Postgrad Med J. 2002;78:286–7. https://doi.org/10.1136/pmj.78.919.286.
Brechmann T, Gunther K, Neid M, Schmiegel W, Tannapfel A. Triggers of histologically suspected drug-induced colitis. World J Gastroenterol. 2019;25:967–79. https://doi.org/10.3748/wjg.v25.i8.967.
Gao H, Peng K, Shi Y, Zhu S, Sun R, Xu C, et al. Development and validation of a novel criterion of histologic healing in ulcerative colitis defined by inflammatory cell enumeration in lamina propria mucosa: A multicenter retrospective cohort in China. Chin Med J. 2024;137:1316–23. https://doi.org/10.1097/CM9.0000000000003154.
Magro F, Gionchetti P, Eliakim R, Ardizzone S, Armuzzi A, Barreiro-de Acosta M, et al. Third European evidence-based consensus on diagnosis and management of ulcerative Colitis. Part 1: Definitions, Diagnosis, Extraintestinal Manifestations, Pregnancy, Cancer Surveillance, Surgery, and Ileo-anal Pouch Disorders. J Crohns Colitis. 2017;11:649–70. https://doi.org/10.1093/ecco-jcc/jjx008.
Gionchetti P, Dignass A, Danese S, Magro Dias FJ, Rogler G, Lakatos PL, et al. 3rd European Evidence-based Consensus on the diagnosis and management of Crohn’s Disease 2016: Part 2: surgical management and special situations. J Crohns Colitis. 2017;11:135–49. https://doi.org/10.1093/ecco-jcc/jjw169.
Zhou G, Yu L, Fang L, Yang W, Yu T, Miao Y, et al. CD177(+) neutrophils as functionally activated neutrophils negatively regulate IBD. Gut. 2018;67:1052–63. https://doi.org/10.1136/gutjnl-2016-313535.
Lu H, Zhang C, Wu W, Chen H, Lin R, Sun R, et al. MCPIP1 restrains mucosal inflammation by orchestrating the intestinal monocyte to macrophage maturation via an ATF3-AP1S2 axis. Gut. 2023;72:882–95. https://doi.org/10.1136/gutjnl-2022-327183.
Ramos GP, Bamidele AO, Klatt EE, Sagstetter MR, Kurdi AT, Hamdan FH, et al. G9a modulates lipid metabolism in CD4 T cells to regulate intestinal inflammation. Gastroenterology. 2023;164:256–271 e210. https://doi.org/10.1053/j.gastro.2022.10.011.
Lin R, Ma C, Fang L, Xu C, Zhang C, Wu X, et al. TOB1 blocks intestinal mucosal inflammation through inducing ID2-mediated suppression of Th1/Th17 cell immune responses in IBD. Cell Mol Gastroenterol Hepatol. 2022;13:1201–21. https://doi.org/10.1016/j.jcmgh.2021.12.007.
Li G, Lin J, Gao X, Su H, Lin R, Gao H, et al. Intestinal epithelial pH-sensing receptor GPR65 maintains mucosal homeostasis by regulating antimicrobial defense and restrains gut inflammation in inflammatory bowel disease. Gut Microbes. 2023;15:2257269. https://doi.org/10.1080/19490976.2023.2257269.
Acknowledgements
We gratefully acknowledge the financial support of the National Natural Science Foundation of China (82370532, 82341219) and Shanghai Hospital Development Center Foundation (SHDC12022118). We thank Professor Youcun Qian (Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, Shanghai) for valuable advice on the experimental design. We also acknowledge the physicians and nurses in the Department of Gastroenterology of Shanghai Tenth People’s Hospital for their assistance with sample collection. We would like to thank BioRender (biorender.com) for providing the tools used to create the scientific illustrations in this manuscript.
Author information
Authors and Affiliations
Contributions
ZL conceived the study. AL, DK, ZF, and HL designed the experimental protocol, conducted the majority of the experiments and statistical analyses, and drafted the manuscript. XG, HG, XW, XL, FK, JL, JH, PY, and XL contributed to data interpretation. JL and ZX provided the clinical specimens. All the authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, A., Kang, D., Feng, Z. et al. YDJC restrains Th1 cell differentiation by blocking SREBP2-mediated cholesterol biosynthesis to alleviate mucosal inflammation in inflammatory bowel disease. Cell Mol Immunol 23, 15–30 (2026). https://doi.org/10.1038/s41423-025-01361-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41423-025-01361-z