Abstract
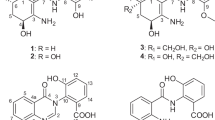
Using genome mining, a new cytotoxic peptide named curacozole was isolated from Streptomyces curacoi. Through ESI-MS and NMR analyses, curacozole was determined to be a macrocyclic peptide containing two isoleucine, two thiazole and three oxazole moieties. Curacozole exhibited potent cytotoxic activity against HCT116 and HOS cancer cells. The proposed biosynthetic gene cluster of curacozole was identified and compared with that of the related compound YM-216391.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Li L, Jiang W, Lu Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol Adv. 2017;35:936–49.
Velasquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol. 2011;15:11–21.
Ju KS, et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc Natl Acad Sci USA. 2015;112:12175–80.
Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–46.
Blin K, et al. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–12.
Blin K, Kazempour D, Wohlleben W, Weber T. Improved lanthipeptide detection and prediction for antiSMASH. PLoS ONE. 2014;9:e89420.
Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43.
Blin K, Medema MH, Kottmann R, Lee SY, Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017;45:D555–9.
Tietz JI, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–8.
Shin-ya K, et al. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123:1262–3.
Kim MY, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc. 2002;124:2098–9.
Amagai K, et al. Identification of a gene cluster for telomestatin biosynthesis and heterologous expression using a specific promoter in a clean host. Sci Rep. 2017;7:3382.
Sohda K, Nagai K, Yamori T, Suzuki K, Tanaka A. YM-216391, a novel cytotoxic cyclic peptide from Streptomyces nobilis. I. fermentation, isolation and biological activities. J Antibiot. 2005;58:27–31.
Sohda K, Hiramoto M, Suzumura K, Takebayashi Y, Suzuki K, Tanaka A. YM-216391, a novel cytotoxic cyclic peptide from Streptomyces nobilis. II. Physico-chemical properties and structure elucidation. J Antibiot. 2005;58:32–6.
Jian XH, et al. Analysis of YM-216391 biosynthetic gene cluster and improvement of the cyclopeptide production in a heterologous host. ACS Chem Biol. 2012;7:646–51.
Kaweewan I, Komaki H, Hemmi H, Kodani S. Isolation and structure determination of new antibacterial peptide curacomycin based on genome mining. Asian J Org Chem. 2017;6:1838–44.
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schaffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–64.
Hosaka T, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27:462–4.
Ochi K, Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol. 2013;97:87–98.
Thong WL, Shin-ya K, Nishiyama M, Kuzuyama T. Methylbenzene-containing polyketides from a Streptomyces that spontaneously acquired rifampicin resistance: structural elucidation and biosynthesis. J Nat Prod. 2016;79:857–64.
Hu H, Zhang Q, Ochi K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J Bacteriol. 2002;184:3984–91.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Harada K, Fujii K, Hayashi K, Suzuki M, Ikai Y, Oka H. Application of D, L-FDLA derivatization to determination of absolute configuration of constituent amino acids in peptide by advanced Marfey’s method. Tetrahedron Lett. 1996;37:3001–4.
Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA. 2010;107:16297–302.
Kodani S, Komaki H, Ishimura S, Hemmi H, Ohnishi-Kameyama M. Isolation and structure determination of a new lantibiotic cinnamycin B from Actinomadura atramentaria based on genome mining. J Ind Microbiol Biotechnol. 2016;43:1159–65.
Kaweewan I, Komaki H, Hemmi H, Kodani S. Isolation and structure determination of a new thiopeptide globimycin from Streptomyces globisporus subsp. globisporus based on genome mining. Tetrahedron Lett. 2018;59:409–14.
Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol. 1996;178:7276–84.
Acknowledgements
This study was supported by the Japan Society for the Promotion of Science by Grants-in-aids (grant number 16K01913). The research was partly supported by the Sasakawa Scientific Research Grant from The Japan Science Society (grant number 2018-3001). The NMR spectra were recorded on Bruker Avance 600 and Avance III HD 800 spectrometers at Advanced Analysis Center, NARO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kaweewan, I., Komaki, H., Hemmi, H. et al. Isolation and structure determination of a new cytotoxic peptide, curacozole, from Streptomyces curacoi based on genome mining. J Antibiot 72, 1–7 (2019). https://doi.org/10.1038/s41429-018-0105-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-018-0105-4
This article is cited by
-
Discovery of anti-Mycobacterium tuberculosis desertomycins from Streptomyces flavofungini TRM90047 based on genome mining and HSQC-TOCSY
Scientific Reports (2024)
-
A putative mechanism underlying secondary metabolite overproduction by Streptomyces strains with a 23S rRNA mutation conferring erythromycin resistance
Applied Microbiology and Biotechnology (2020)
-
Lincomycin-Induced Secondary Metabolism in Streptomyces lividans 66 with a Mutation in the Gene Encoding the RNA Polymerase Beta Subunit
Current Microbiology (2020)