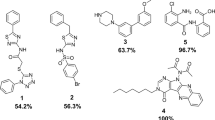
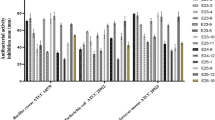
Abstract
Staphylococcus aureus is one of the most common nosocomial biofilm-forming pathogens worldwide that has developed resistance mechanisms against majority of the antibiotics. Therefore, the search of novel antistaphylococcal agents with unexploited mechanisms of action, especially with antibiofilm activity, is of great interest. Seryl-tRNA synthetase is recognized as a promising drug target for the development of antibacterials. We have carried out molecular docking of compounds with antistaphycoccal activity, which were earlier found by us using phenotypic screening, into synthetic site of S. aureus SerRS and found seven hit compounds with low inhibitory activity. Further, we have performed search of S. aureus SerRS inhibitors among compounds which were previously tested by us for inhibitory activity toward S. aureus ThrRS, that belong to the same class of aminoacyl-tRNA synthetases. Among them six hits were identified. We have selected four compounds for antibacterial study and found that the most active compound 1-methyl-3-(1H-imidazol-1-methyl-2-yl)-5-nitro-1H-indazole has MIC values toward S. aureus multidrug-resistant clinical isolates ranging from 78.12 to 156.2 µg/ml. However, this compound precipitated during anti-biofilm study. Therefore, we used 3-[N’-(2-hydroxy-3-methoxybenzylidene)hydrazino]-6-methyl-4H-[1,2,4]triazin-5-one with better solubility (ClogS value = 2.9) among investigated compounds toward SerRS for anti-biofilm study. It was found that this compound has a significant inhibitory effect on the growth of planktonic and biofilm culture of S. aureus 25923 with MIC value of 32 µg ml−1. At the same time, this compound does not reveal antibacterial activity toward Esherichia coli ATCC 47076. Therefore, this compound can be proposed as effective antiseptic toward multidrug-resistant biofilm-forming S. aureus isolates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Berge A, Carlsén C, Petropoulos A, Gadler F, Rasmussen M. Staphylococcus aureus bacteraemia, cardiac implantable electronic device, and the risk of endocarditis: a retrospective population-based cohort study. Eur J Clin Microbiol Infect Dis. 2023;42:583–91.
García de la Mària C, et al. Emerging issues on Staphylococcus aureus endocarditis and the role in therapy of daptomycin plus fosfomycin. Expert Rev Anti Infect Ther. 2023;21:281–93.
Tournaye E, Hollering P, De Roover D, Dossche K, Vercauteren SRW. Staphylococcus aureus sepsis and hemoptysis as messengers of a rather impractically located mycotic aneurysm. Acta Chir Belg. 2023;123:430–35.
Chen Y, et al. The effect of Staphylococcus aureus on innate and adaptive immunity and potential immunotherapy for S. aureus-induced osteomyelitis. Front Immunol. 2023;14:1219895.
van Soest TM, et al. Community-acquired Staphylococcus aureus meningitis in adults. J Infect. 2023;86:239–44.
Venkateswaran P, et al. Revisiting ESKAPE Pathogens: virulence, resistance, and combating strategies focusing on quorum sensing. Front Cell Infect Microbiol. 2023;13:1159798.
Gherardi G. Staphylococcus aureus Infection: Pathogenesis and antimicrobial resistance. Int J Mol Sci. 2023;24:8182.
Huynh TQ, et al. Genomic alterations involved in fluoroquinolone resistance development in Staphylococcus aureus. PLoS One. 2023;18:e0287973.
Ippolito G, Leone S, Lauria FN, Nicastri E, Wenzel RP. Methicillin-resistant Staphylococcus aureus:the superbug. Int J Infect Dis. 2010;14:S7–S11.
Kaur DC, Chate SS. Study of antibiotic resistance pattern in methicillin-resistant Staphylococcus aureus with special reference to newer antibiotic. J Glob Infect Dis. 2015;7:78–84.
Arunkumar V, Prabagaravarthanan R, Bhaskar M. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infections among patients admitted in critical care units in a tertiary care hospital. Int J Res Med Sci. 2017;5:2362–66.
McGuinness WA, Malachowa N, DeLeo FR. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90:269–81.
García-Angulo VA, et al. Isolation and first draft genome sequence of a linezolid-dependent Staphylococcus aureus clinical strain. Future Microbiol. 2020;15:1123–29.
Yoo IY, Kang OK, Shim HJ, Huh HJ, Lee NY. Linezolid resistance in methicillin-resistant Staphylococcus aureus in Korea: high rate of false resistance to linezolid by the VITEK 2 system. Ann Lab Med. 2020;40:57–62.
Tran NN, Morrisette T, Jorgensen SCJ, Orench-Benvenutti JM, Kebriaei R. Current therapies and challenges for the treatment of Staphylococcus aureus biofilm-related infections. Pharmacotherapy. 2023;43:816–32.
Pang L, Weeks SD, Van Aerschot A. Aminoacyl-tRNA synthetases as valuable targets for antimicrobial drug discovery. Int J Mol Sci. 2021;22:1750.
Francklyn CS, Mullen P. Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem. 2019;294:5365–85.
Zeng Y, Roy H, Patil PB, Ibba M, Chen S. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob Agents Chemother. 2009;53:4619–27.
Battenberg OA, Yang Y, Verhelst SH, Sieber SA. Target profiling of 4-hydroxyderricin in S. aureus reveals seryl-tRNA synthetase binding and inhibition by covalent modification. Mol Biosyst. 2013;9:343–51.
Cain R, et al. Structure-guided enhancement of selectivity of chemical probe inhibitors targeting bacterial seryl-tRNA synthetase. J Med Chem. 2019;62:9703–17.
Bodian DL, et al. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–78.
Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput-Aided Mol Des. 2001;15:411–28.
Ring CS, et al. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc Natl Acad Sci USA. 1993;90:3583–7.
Stoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–50.
Yakovenko OY, Oliferenko A, Golub A, Bdzhola V, Yarmoluk S. The new method of distribution integrals evaluations for high throughput virtual screening. Ukr Bioorg Acta. 2007;1:52–62.
Yakovenko O, Oliferenko AA, Bdzhola VG, Palyulin VA, Zefirov NS. Kirchhoff atomic charges fitted to multipole moments: implementation for a virtual scrrening system. J Comput Chem. 2008;29:1332–43.
Kovalenko OP, et al. Dual-target inhibitors of mycobacterial aminoacyl-tRNA synthetases among N-benzylidene-N’-thiazol-2-yl-hydrazines. Medchemcomm. 2019;10:2161–9.
Discovery Studio Visualizer 4.0. https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php, (accessed May 2019).
Studier F. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34.
Volynets G, et al. Identification of novel antistaphylococcal hit compounds targeting sortase A. Molecules. 2021;26:7095.
Rybak MY, et al. Rational design of hit compounds targeting Staphylococcus aureus threonyl-tRNA synthetase. ACS Omega. 2021;6:24910–8.
EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance, Version 1.0, December 2013. EUCAST; Växjö, Sweden: 2013.
Acknowledgements
This work was supported by the NAS of the Ukraine grant № 0120U000079. The authors are thankful to Prof. David Roper (School of Life Sciences, University of Warwick, United Kingdom) for the gift of plasmid encoding S. aureus SerRS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Volynets, G.P., Iungin, O.S., Gudzera, O.I. et al. Identification of novel antistaphylococcal hit compounds. J Antibiot 77, 665–678 (2024). https://doi.org/10.1038/s41429-024-00752-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-024-00752-0