Abstract
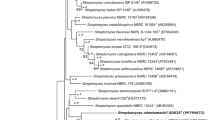
A novel actinomycete, designated as TPMA0078T, was isolated from a soil sample collected in Shinjuku, Tokyo, Japan. 16S rRNA gene sequence analysis indicated that strain TPMA0078T belongs to the genus Actinoplanes and is closely related to Actinoplanes regularis IFO 12514T (99.86% 16S rRNA gene sequence similarity). The spores of strain TPMA0078T were motile, and the sporangia were cylindrical. The diamino acids in the cell wall peptidoglycan of strain TPMA0078T were meso-diaminopimelic acid and 3OH-meso-diaminopimelic acid. Whole-cell sugars were glucose and mannose, with galactose as a minor component. The major cellular fatty acids identified were iso-C15:0, iso-C16:0, and anteiso-C17:0. The predominant menaquinone was MK-9(H4), and the principal polar lipid was phosphatidylethanolamine. These chemotaxonomic properties of strain TPMA0078T were consistent with those of Actinoplanes. Meanwhile, digital DNA–DNA hybridization and average nucleotide identity values showed low relatedness between strain TPMA0078T and A. regularis NBRC 12514T. Furthermore, several phenotypic properties of strain TPMA0078T distinguished it from those of closely related species. Based on its genotypic and phenotypic characteristics, strain TPMA0078T represents a novel species of the genus Actinoplanes, for which the name Actinoplanes kirromycinicus sp. nov. is proposed. The type strain is TPMA0078T (=NBRC 116422T = TBRC 18262T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Couch JN. Actinoplanes, a new genus of the Actinomycetales. J Elisha Mitchell Sci Soc. 1950;66:87–92.
Stackebrandt E, Kroppenstedt RM. Union of the genera Actinoplanes couch, Ampullariella couch, and Amorphosporangium couch in a redefined genus Actinoplanes. Syst Appl Microbiol. 1987;9:110–4.
Marcone GL, et al. Classification of Actinoplanes sp. ATCC 33076, an actinomycete that produces the glycolipodepsipeptide antibiotic ramoplanin, as Actinoplanes ramoplaninifer sp. nov. Int J Syst Evol Microbiol. 2017;67:4181–8.
Habib N, et al. Actinoplanes deserti sp. nov., isolated from a desert soil sample. Antonie Van Leeuwenhoek. 2018;111:2303–10.
Luo X, et al. Actinoplanes flavus sp. nov., a novel cellulase-producing actinobacterium isolated from coconut palm rhizosphere soil. Int J Syst Evol Microbiol. 2021;71:004990.
Ding L-M, et al. Three novel Actinoplanes species isolated by using polyaspartic acid as a water-retaining agent for the enrichment in situ. Int J Syst Evol Microbiol. 2023;73:005705.
Goodfellow M, Stanton LJ, Simpson K, Minnikin DE. Numerical and chemical classification of Actinoplanes and some related actinomycetes. J Gen Microbiol. 1990;136:19–36.
Tamura T, Hatano K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int J Syst Evol Microbiol. 2001;51:2119–25.
Sazak A, Sahin N, Camas M. Actinoplanes abujensis sp. nov., isolated from Nigerian arid soil. Int J Syst Evol Microbiol. 2012;62:960–5.
Bardone MR, Paternoster M, Coronelli C. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J Antibiot. 1978;31:170–7.
Debono M, et al. Actaplanin, new glycopeptide antibiotics produced by Actinoplanes missouriensis. The isolation and preliminary chemical characterization of actaplanin. J Antibiot. 1984;37:85–95.
Wagman GH, et al. New polyene antifungal antibiotic produced by a species of Actinoplanes. Antimicrob Agents Chemother. 1975;7:457–61.
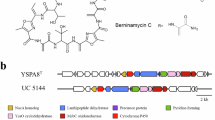
Weber T, et al. Molecular analysis of the kirromycin biosynthetic gene cluster revealed β-alanine as precursor of the pyridone moiety. Chem Biol. 2008;15:175–88.
Wolf H, Chinali G, Parmeggiani A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc Natl Acad Sci USA 1974;71:4910–4.
Beretta G, Le Monnier F, Selva E, Marinelli F. A novel producer of the antibiotic kirromycin belonging to the genus Actinoplanes. J Antibiot. 1993;46:1175–7.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Wakisaka Y, Kawamura Y, Yasuda Y, Koizumi K, Nishimoto Y. A selective isolation procedure for Micromonospora. J Antibiot. 1982;35:822–36.
Saito H, Miura KI. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–29.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. J Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–16.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–204.
Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963.
Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9.
Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:36–41.
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60.
Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–6.
Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182.
Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–800.
Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for selective isolation of soil actinomycetes. J Ferment Technol. 1987;65:501–9.
Uchida K, et al. Characterization of Actinoplanes missouriensis spore flagella. Appl Environ Microbiol. 2011;77:2559–62.
Kammanee S, et al. Saccharopolyspora oryzae sp. nov., isolated from rhizosphere soil of the wild rice species Oryza rufipogon. J Antibiot. 2023;76:658–64.
Pridham TG, Gottlieb D. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J Bacteriol. 1948;56:107–14.
Hamada M, et al. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J Antibiot. 2012;65:427–31.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–31.
Minnikin DE, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–41.
Sasser M. MIDI technical note 101. Identification of bacteria by gas chromatography of cellular fatty acids. Newark: DE MIDI inc; 1990.
Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 2005;102:2567–72.
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 2009;106:19126–31.
Chun J, Rainey FA. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol. 2014;64:316–24.
Robertsen HL, et al. Filling the gaps in the kirromycin biosynthesis: deciphering the role of genes involved in ethylmalonyl-CoA supply and tailoring reactions. Sci Rep. 2018;8:3230.
Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Letter: roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot. 1974;27:86–7.
Awakawa T, et al. Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. Chembiochem. 2011;12:439–48.
Yim G, et al. Harnessing the synthetic capabilities of glycopeptide antibiotic tailoring enzymes: characterization of the UK-68,597 biosynthetic cluster. Chembiochem. 2014;15:2613–23.
Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–43.
Funding
This study was supported in part by Grants-in-Aid for the Toho University Grant for Research Initiative Program (TUGRIP) from Toho University (to YI and YA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, J., Iizaka, Y., Hamada, M. et al. Actinoplanes kirromycinicus sp. nov., isolated from soil. J Antibiot 77, 657–664 (2024). https://doi.org/10.1038/s41429-024-00756-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-024-00756-w