Abstract
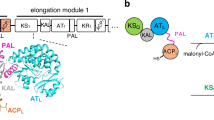
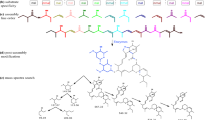
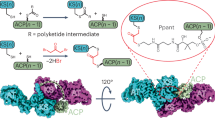
Genome mining is a powerful strategy for finding biosynthetic gene clusters (BGCs) for unprecedented natural products and their biosynthetic mechanisms. In this study, to obtain novel type polyketides, we performed genome mining focusing on three perspectives for novelty: 1) amino acid sequence-based classification, 2) unique domain architectures of polyketide synthase, and 3) predicted structural features from protein modeling. As a result, we discovered a BGC consisting of a highly reducing polyketide synthase and a non-reducing polyketide synthase (NR-PKS), which harbors noncanonical tandem acyl carrier protein (ACP) domains and a structurally characteristic thioesterase (TE) domain. Heterologous expression revealed that the cyrl cluster produces a novel dimer of long-chain alkylresorcinolic acid (1). Furthermore, site-directed mutagenesis of each ACP domain revealed that both domains are essential for efficient dimerization. This study provides the first example of a fungal dimer-forming NR-PKS in which the tandem ACP domains work “in-series” with nonredundant roles to construct a dimeric long-chain alkylresorcinolic acid. The in silico analysis suggested that the TE domain led the substrate to a dimerizable form. Since 1 is structurally related to integracins with HIV-1 integrase inhibition, our findings provide insight into integracins biosynthesis and offer a basis for generating new integracin derivatives.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26.
Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot. 2012;65:385–95.
Rokas A, Mead ME, Steenwyk JL, Raja HA, Oberlies NH. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat Prod Rep. 2020;37:868–78.
Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91.
Vesth TC, Nybo JL, Theobald S, Frisvad JC, Larsen TO, Nielsen KF, et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat Genet. 2018;50:1688–95.
Kaneko A, Morishita Y, Tsukada K, Taniguchi T, Asai T. Post-genomic approach-based discovery of alkylresorcinols from a cricket-associated fungus, Penicillium soppi. Org Biomol Chem. 2019;5239–43.
Morishita Y, Zhang H, Taniguchi T, Mori K, Asai T. The discovery of fungal polyene macrolides via a postgenomic approach reveals a polyketide macrocyclization by trans-acting thioesterase in fungi. Org Lett. 2019;21:4788–92.
Tsukada K, Shinki S, Kaneko A, Murakami K, Irie K, Murai M, et al. Synthetic biology based construction of biological activity-related library of fungal decalin-containing diterpenoid pyrones. Nat Commun. 2020;11:1830.
Morishita Y, Aoki Y, Ito M, Hagiwara D, Torimaru K, Morita D, et al. Genome mining-based discovery of fungal macrolides modified by glycosylphosphatidylinositol (GPI)-ethanolamine phosphate transferase homologues. Org Lett. 2020;22:5876–9.
Homma Y, Sugawara A, Morishita Y, Tsukada K, Ozaki T, Asai T. Discovery of a cyclic depsipeptide from Chaetomium mollipilium by the genome mining approach. Org Lett. 2022;24:3504–9.
Sato Y, Morishita Y, Homma Y, Sugawara A, Moussa AY, Elissawy AM, et al. Biosynthesis of circumdatins employs an anthranilate tailoring pathway for NRPS substrate supplies. Org Lett. 2025;27:7170–5.
Sato F, Sonohara T, Fujiki S, Sugawara A, Morishita Y, Ozaki T, et al. Genome mining of labdane-related diterpenoids: discovery of the two-enzyme pathway leading to (−)-sandaracopimaradiene in the fungus Arthrinium sacchari. Beilstein J Org Chem. 2024;20:714–20.
Shi Y, Ozaki T, Sugawara A, Morishita Y, Pei Tan Y, Shivas RG, et al. A new 3,6-dialkyl-α-pyrone produced by the heterologous expression of a PKS-NRPS hybrid enzyme derived from a Pestalotiopsis endophyte. Tetrahedron Lett. 2024;134:154865.
Hirokawa M, Ozaki T, Tsukada K, Sugawara A, Morishita Y, Asai T. Ketosynthase domain catalyzes β-lactonization in the biosynthesis of the HMG-CoA synthase inhibitor hymeglusin. J Am Chem Soc. 2025;147:25136–41.
Shi Y, Ozaki T, Morishita Y, Taniguchi T, Sato H, Aoyama Y, et al. Genome mining-based discovery of pyrano[2,3-c] pyrrole type natural products possessing alkyl side chain with branched methyl groups. Org Lett. 2025;27:8580–5.
Furumura S, Ozaki T, Sugawara A, Morishita Y, Tsukada K, Ikuta T, et al. Identification and functional characterization of fungal chalcone synthase and chalcone isomerase. J Nat Prod. 2023;86:398–405.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Abe T, Shiratori H, Kashiwazaki K, Hiasa K, Ueda D, Taniguchi T, et al. Structural-model-based genome mining can efficiently discover novel non-canonical terpene synthases hidden in genomes of diverse species. Chem Sci. 2024;15:10402–7.
Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol. 2017;13:895–901.
Harvey CJB, Tang M, Schlecht U, Horecka J, Fischer CR, Lin HC, et al. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci Adv. 2018;4:eaar5459.
Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: from genes to molecules. J Org Chem. 2012;77:9933–53.
Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–89.
Chen L, Wei X, Matsuda Y. Depside bond formation by the starter-unit acyltransferase domain of a fungal polyketide synthase. J Am Chem Soc. 2022;144:19225–30.
Ji Q, Xiang H, Wang WG, Matsuda Y. Mechanism behind the programmed biosynthesis of heterotrimerifc fungdal depstide thielavin A. Angew Chem Int Ed. 2024;63:e202402663.
Skellam E. Biosynthesis of fungal polyketides by collaborating and trans-acting enzymes. Nat Prod Rep. 2022;39:754–83.
Gaffoor I, Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol. 2006;72:1793–9.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9.
Reeves CD, Hu Z, Reid R, Kealey JT. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl Environ Microbiol. 2008;74:5121–9.
Maruyama JI, Kitamoto K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae. Biotechnol Lett. 2008;30:1811–7.
Ohtani I, Kusumi T, Kashman Y, Kakisawa H. High-Field FT NMR application of Mosher’s method. the absolute configurations of marine terpenoids. J Am Chem Soc. 1991;113:4092–6.
Singh SB, Zink DL, Bills GF, Pelaez F, Teran A, Collado J, et al. Discovery, structure and HIV-1 integrase inhibitory activities of integracins, novel dimeric alkyl aromatics from Cytonaema sp. Tetrahedron Lett. 2002;43:1617–20.
Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–97.
Wang S, Xu Y, Maine EA, Wijeratne EMK, Espinosa-Artiles P, Gunatilaka AAL, et al. Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol. 2008;15:1328–38.
Xu Y, Zhou T, Espinosa-Artiles P, Tang Y, Zhan J, Molnár I. Insights into the biosynthesis of 12-membered resorcylic acid lactones from heterologous production in Saccharomyces cerevisiae. ACS Chem Biol. 2014;9:1119–27.
Xu Y, Espinosa-Artiles P, Schubert V, Xu Yming, Zhang W, Lin M, et al. Characterization of the biosynthetic genes for 10,11- dehydrocurvularin, a heat shock response-modulating anticancer fungal polyketide from Aspergillus terreus. Appl Environ Microbiol. 2013;79:2038–47.
Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9.
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–4.
Wang J, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu S, et al. The conserved domain database in 2023. Nucleic Acids Res. 2023;51:D384–8.
Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–44.
Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Chen EA, Porter LL. SSDraw: Software for generating comparative protein secondary structure diagrams. Protein Sci. 2023;32:e4836.
Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91.
Tosco P, Stiefl N, Landrum G. Bringing the MMFF force field to the RDKit: implementation and validation. J Cheminf. 2014;6:37.
Acknowledgements
The authors are grateful to the Experimental Station for Medicinal Plant Studies (Graduate School of Pharmaceutical Sciences, Tohoku University) for the isolation of Cytospora sp. We thank Prof. Katsuya Gomi (Tohoku University) and Prof. Katsuhiko Kitamoto (The University of Tokyo) for providing the expression vectors and the fungal strain, Prof. Hideaki Oikawa (Hokkaido University), Prof. Atsushi Minami (Institute of Science Tokyo), and Prof. Jun-ichi Maruyama (The University of Tokyo) for the genome editing system of A. oryzae. This work was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grants JP23K24038 (to TA), JP23KJ0176 (to YH), and JP24K18294 (to YM), JST-ALCANext Japan grant number JPMJAN23D4 (to TA) from Japan Science and Technology Agency, Advanced Research & Development Programs for Medical Innovation (AMED-CREST) grant JP22gm1610007 (to TA) from the Japan Agency for Medical Research and Development, and the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED [under grants JP24am121038 (to TA) and JP24ama121040 (to TO)].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Homma, Y., Hasegawa, T., Morishita, Y. et al. Genome mining-based discovery of an atypical fungal non-reducing polyketide synthase encoding dimeric alkylresorcinol biosynthesis. J Antibiot (2026). https://doi.org/10.1038/s41429-025-00891-y
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41429-025-00891-y